|
Size: 1185
Comment:
|
← Revision 21 as of 2022-08-05 14:31:35 ⇥
Size: 1956
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from Automated Defacing Tools ## page was renamed from mri_deface |
|
| Line 2: | Line 4: |
| [[https://surfer.nmr.mgh.harvard.edu/fswiki/MiDeFace|See MiDeFace, our newest tool for defacing]] |
|
| Line 6: | Line 10: |
| Cite this paper is using mri_deface: [[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2408762/|A Technique for the Deidentification of Structural Brain MR Images]] |
|
| Line 7: | Line 13: |
| * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface-v1.22-Linux64.gz|mri_deface v1.22 for Linux 64bit]] * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface-v1.22-Linux.gz|mri_deface v1.22 for Linux]] * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface-v1.22-MacOS-Leopard-intel.gz|mri_deface v1.22 for Mac OSX Leopard Intel]] |
* [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface_linux|mri_deface v1.22 for Linux]] * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface_osx|mri_deface v1.22 for Mac OS]] |
| Line 12: | Line 18: |
| Line 13: | Line 20: |
| Line 14: | Line 22: |
| gunzip mri_deface-v1.22-Linux64.gz cp mri_deface-v1.22-Linux64.gz mri_deface |
mv mri_deface_linux mri_deface |
| Line 20: | Line 27: |
| Then, example usage, using your orig.mgz: | The command usage is: |
| Line 22: | Line 31: |
| ./mri_deface orig.mgz talairach_mixed_with_skull.gca face.gca orig_defaced.mgz | ./mri_deface <input> talairach_mixed_with_skull.gca face.gca <output> |
| Line 24: | Line 33: |
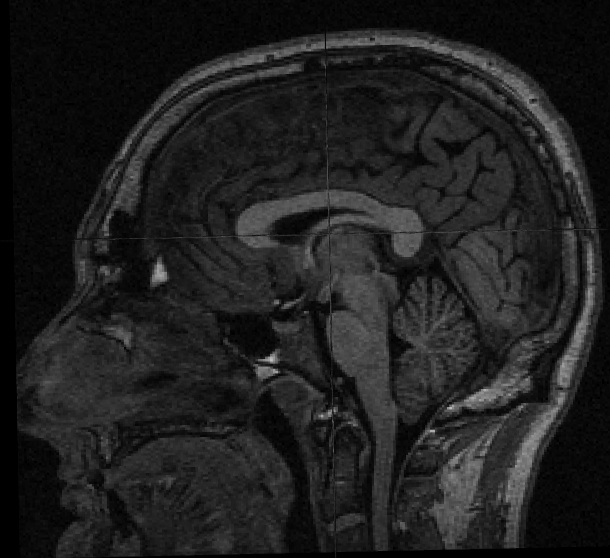
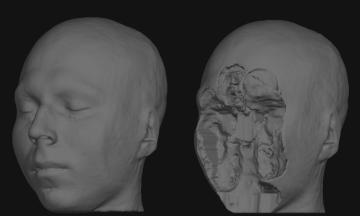
So as an example, to deface your T1-weighted input file, which can be a dicom or nifti, run: {{{ ./mri_deface T1.nii.gz talairach_mixed_with_skull.gca face.gca T1_defaced.nii.gz }}} You will need a volume viewer to view the resulting defaced file. The freesurfer package includes the 'freeview' viewer, but there are many others capable of reading dicom and nifti volumes. [[http://www.mccauslandcenter.sc.edu/mricro/mricron/|MRIcron]] is a good option (use MRIcron, not MRIcro, because you will want nifti support). Sample input before and after defacing: {{attachment:sample_T1_input.jpg}} {{attachment:sample_T1_input_defaced.jpg}} |
Automated Defacing Tools
See MiDeFace, our newest tool for defacing
See also MBIRN page
Cite this paper is using mri_deface: A Technique for the Deidentification of Structural Brain MR Images
Note: A license file is no longer necessary to use these tools:
Use gunzip to decompress these downloads, ie.:
mv mri_deface_linux mri_deface chmod a+x mri_deface gunzip talairach_mixed_with_skull.gca.gz gunzip face.gca.gz
The command usage is:
./mri_deface <input> talairach_mixed_with_skull.gca face.gca <output>
So as an example, to deface your T1-weighted input file, which can be a dicom or nifti, run:
./mri_deface T1.nii.gz talairach_mixed_with_skull.gca face.gca T1_defaced.nii.gz
You will need a volume viewer to view the resulting defaced file. The freesurfer package includes the 'freeview' viewer, but there are many others capable of reading dicom and nifti volumes. MRIcron is a good option (use MRIcron, not MRIcro, because you will want nifti support).
Sample input before and after defacing: