|
Size: 1514
Comment:
|
Size: 1926
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from Automated Defacing Tools ## page was renamed from mri_deface |
|
| Line 2: | Line 4: |
| [[https://surfer.nmr.mgh.harvard.edu/fswiki/MiDeFace|See MiDeFace]] |
|
| Line 6: | Line 10: |
| Cite this paper is using mri_deface: [[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2408762/|A Technique for the Deidentification of Structural Brain MR Images]] |
|
| Line 7: | Line 13: |
| * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface-v1.22-Linux64.gz|mri_deface v1.22 for Linux 64bit]] * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface-v1.22-Linux.gz|mri_deface v1.22 for Linux]] * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface-v1.22-MacOS-Leopard-intel.gz|mri_deface v1.22 for Mac OSX Leopard Intel]] |
* [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface_linux|mri_deface v1.22 for Linux]] * [[https://surfer.nmr.mgh.harvard.edu/pub/dist/mri_deface/mri_deface_osx|mri_deface v1.22 for Mac OS]] |
| Line 12: | Line 18: |
| Line 13: | Line 20: |
| Line 14: | Line 22: |
| gunzip mri_deface-v1.22-Linux64.gz cp mri_deface-v1.22-Linux64.gz mri_deface |
mv mri_deface_linux mri_deface |
| Line 20: | Line 27: |
| Then, example usage, using your orig.mgz: | The command usage is: |
| Line 22: | Line 31: |
| ./mri_deface orig.mgz talairach_mixed_with_skull.gca face.gca orig_defaced.mgz | ./mri_deface <input> talairach_mixed_with_skull.gca face.gca <output> |
| Line 25: | Line 34: |
| If you do not have the 'orig.mgz' file as input (which would only be the case if you have processed your data using the Freesurfer package), then just substitute the name of your T1-weighted input file, which can be a dicom or nifti: | So as an example, to deface your T1-weighted input file, which can be a dicom or nifti, run: |
| Line 27: | Line 37: |
| ./mri_deface my_T1.nii talairach_mixed_with_skull.gca face.gca my_T1_defaced.nii | ./mri_deface T1.nii.gz talairach_mixed_with_skull.gca face.gca T1_defaced.nii.gz |
| Line 29: | Line 39: |
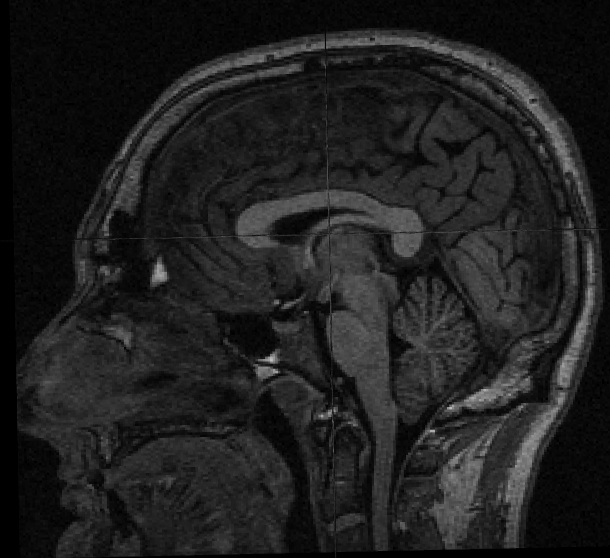
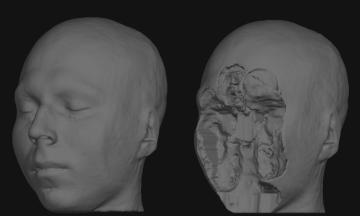
You will need a volume viewer to view the resulting defaced file. The freesurfer package includes the 'freeview' viewer, but there are many others capable of reading dicom and nifti volumes. [[http://www.mccauslandcenter.sc.edu/mricro/mricron/|MRIcron]] is a good option (use MRIcron, not MRIcro, because you will want nifti support). Sample input before and after defacing: {{attachment:sample_T1_input.jpg}} {{attachment:sample_T1_input_defaced.jpg}} |
Automated Defacing Tools
See also MBIRN page
Cite this paper is using mri_deface: A Technique for the Deidentification of Structural Brain MR Images
Note: A license file is no longer necessary to use these tools:
Use gunzip to decompress these downloads, ie.:
mv mri_deface_linux mri_deface chmod a+x mri_deface gunzip talairach_mixed_with_skull.gca.gz gunzip face.gca.gz
The command usage is:
./mri_deface <input> talairach_mixed_with_skull.gca face.gca <output>
So as an example, to deface your T1-weighted input file, which can be a dicom or nifti, run:
./mri_deface T1.nii.gz talairach_mixed_with_skull.gca face.gca T1_defaced.nii.gz
You will need a volume viewer to view the resulting defaced file. The freesurfer package includes the 'freeview' viewer, but there are many others capable of reading dicom and nifti volumes. MRIcron is a good option (use MRIcron, not MRIcro, because you will want nifti support).
Sample input before and after defacing: