|
Size: 9790
Comment:
|
Size: 3670
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from BrainmapOntology_Yeo2014 | |
| Line 3: | Line 4: |
| = Cortical Parcellation Estimated by Intrinsic Functional Connectivity = Data from 1000 young, healthy adults were registered using surface-based alignment. All data were acquired on Siemens 3T scanners using the same functional and structural sequences. A clustering approach was employed to identify and replicate networks of functionally coupled regions across the cerebral cortex. The results revealed local networks confined to sensory and motor cortices as well as distributed networks of association regions that form interdigitated circuits. Within the sensory and motor cortices, functional connectivity followed topographic representations across adjacent areas. In association cortex, the connectivity patterns often showed abrupt transitions between network boundaries, forming largely parallel circuits. The parcellations of the cerebral cortex into 7 and 17 networks in FreeSurfer surface space and nonlinear MNI152 are available for download. |
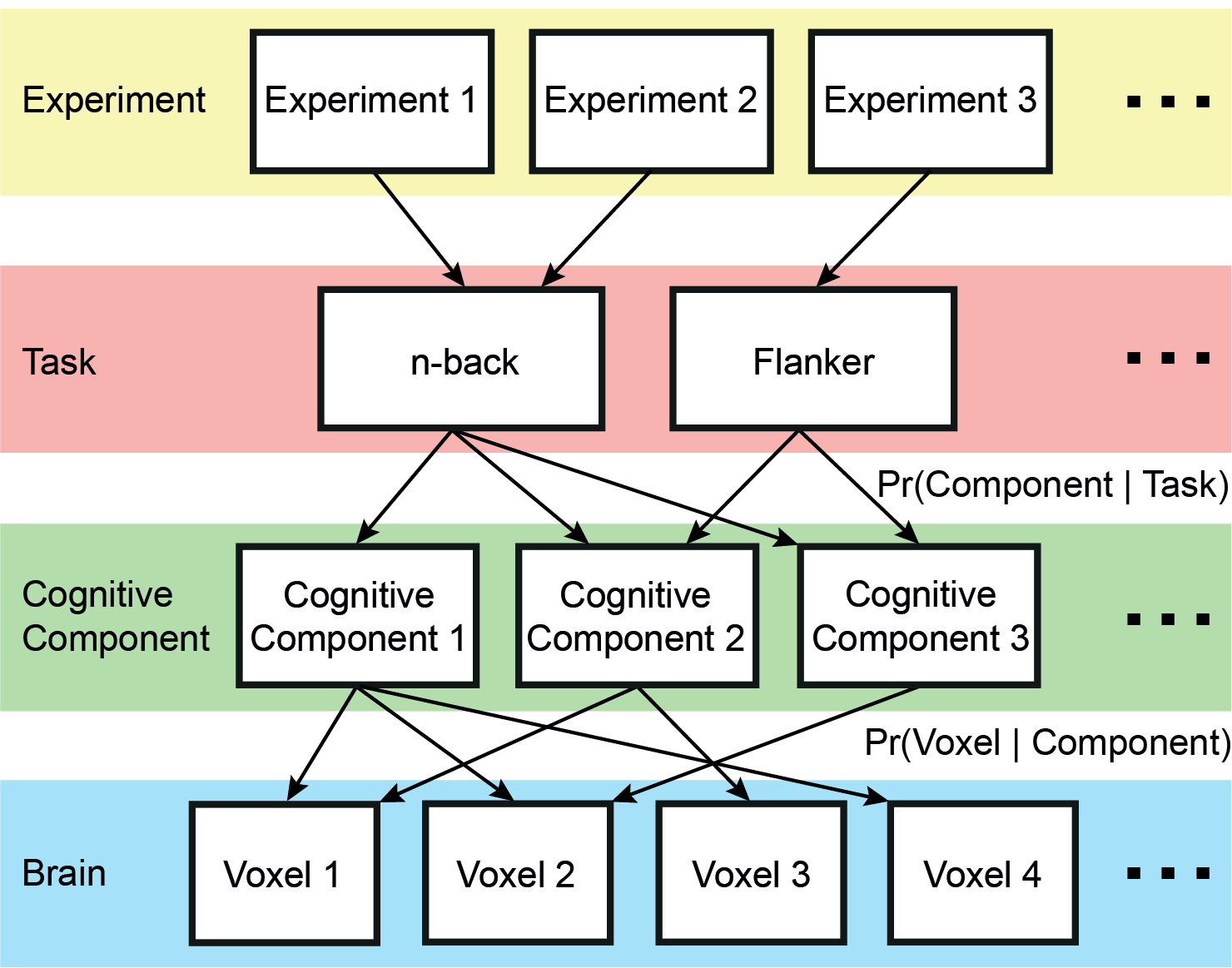
= Nested Cognitive Ontology Estimated From 10,449 Experiments = ||We applied a hierarchical Bayesian model (figure on right) on 10,449 Brainmap experiments across 83 behavioral tasks. By formalizing the notion that performing a task engages multiple cognitive components, each supported by overlapping brain regions, we identified cognitive components that are shared across tasks. Components divided into subcomponents as the number of estimated components increased, revealing a nested ontology. The components enabled the derivation of quantitative maps of functional specialization, revealing complex zones of frontal and parietal regions ranging from being highly specialized to highly flexible. The network organization of the specialized and flexible regions was explored with an independent resting-state fMRI dataset (N=1000). Cortical regions specialized for the same components were strongly coupled, suggesting that components function as partially isolated networks. Functionally flexible regions participate in multiple components to different degrees. This heterogeneous selectivity was strongly predicted by the connectivity between flexible and specialized regions. Functionally flexible regions might support the core of the brain’s information processing capacity, binding or integrating the processing power of segregated, specialized brain networks. The estimated components in FreeSurfer fsaverage space and MNI152 space, as well as specificity and flexibility estimates are available for download. ||{{attachment:BrainmapCognitiveOntology_Yeo2014_model.png|Hierarchical Bayesian Model| width=400 height=233}}|| |
| Line 7: | Line 9: |
| [[http://www.ncbi.nlm.nih.gov/pubmed/21653723|Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L., Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106(3):1125-65, 2011.]] | [[ |Yeo BTT, Krienen FM, Eickhoff SB, Yaakub SN, Fox PT, Buckner RL, Asplund CL, Chee MWL. Functional Specialization and Flexibility in Human Association Cortex. Submitted.]] |
| Line 9: | Line 11: |
| == Parcellations in FreeSurfer Surface Space == ||'''7 Network Estimate''' ||'''7 Network Confidence'''||'''17 Network Estimate''' ||'''17 Network Confidence'''|| || {{attachment:Yeo2011_JNeurophysiol_7networks_lateral.png}} || {{attachment:Yeo2011_JNeurophysiol_7networks_lateral_confidence.png}}|| {{attachment:Yeo2011_JNeurophysiol_17networks_lateral.png}} || {{attachment:Yeo2011_JNeurophysiol_17networks_lateral_confidence.png}}|| || {{attachment:Yeo2011_JNeurophysiol_7networks_medial.png}} || {{attachment:Yeo2011_JNeurophysiol_7networks_medial_confidence.png}}|| {{attachment:Yeo2011_JNeurophysiol_17networks_medial.png}} || {{attachment:Yeo2011_JNeurophysiol_17networks_medial_confidence.png}}|| |
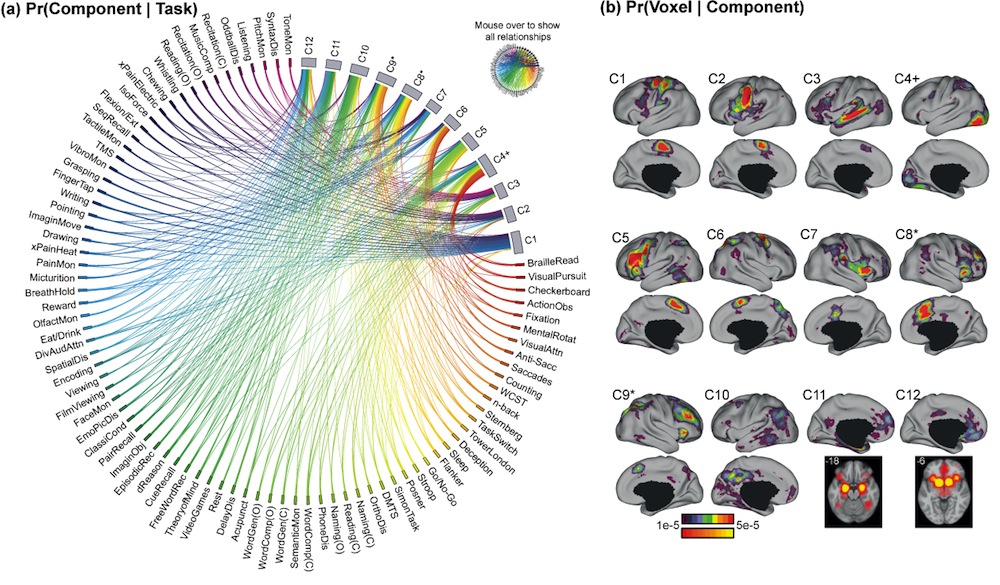
== Interactive 12-Component Cognitive Ontology == [[http://www.ece.nus.edu.sg/stfpage/ybtt/papers/2014Brainmap/Interactive/index.html|Interactive 12-component Ontology can be accessed here!]] We found the interactive version of the static ontology (below) extremely useful. |
| Line 14: | Line 14: |
| ==== Downloads ==== [[ftp://surfer.nmr.mgh.harvard.edu/pub/data/Yeo_JNeurophysiol11_FreeSurfer.zip|Resting State Cortical Parcellation in fsaverage, fsaverage6 and fsaverage5 Space]]. These subjects are also currently in the developmental version of FreeSurfer which can be accessed from the Martinos center network (/autofs/cluster/freesurfer/centos4.0_x86_64/dev/subjects/). These subjects will be officially included in some future FreeSurfer release. Note however, the zip file linked here corresponds exactly to that from the reference, while functional parcellations in the developmental and official release versions of FreeSurfer are subjected to modifications/improvements. |
<<BR>> |
| Line 17: | Line 16: |
| ==== Information about Downloads ==== There are three folders in "`Yeo_JNeurophysiol11_FreeSurfer.zip`", corresponding to the "fsaverage", "fsaverage5" and "fsaverage6" surface space. "fsaverage" contains the high resolution version of the parcellation, while "fsaverage6" and "fsaverage5" contain lower resolution versions of the parcellation. The parcellations were computed in "fsaverage5" space and upsampled to "fsaverage6" and "fsaverage". |
[[http://www.ece.nus.edu.sg/stfpage/ybtt/papers/2014Brainmap/Interactive/index.html|{{attachment:BrainmapCognitiveOntology_Yeo2014_12ComponentOverview.jpg}}|target="_blank"]] |
| Line 20: | Line 18: |
| The structure of each folder follows that of a preprocessed freesurfer subject. In particular, "fsaverage/label/", "fsaverage5/label/", "fsaverage6/label/" contain all the parcellation and confidence files. For example, "fsaverage/label/rh.Yeo2011_7Networks_N1000.annot" is the 7-network parcellation for 1000 subjects on the right hemisphere and "fsaverage/label/lh.Yeo2011_17NetworksConfidence_N1000.mgz" is the confidence map for the 17-network parcellation for 1000 subjects on the left hemisphere. | == Downloads == [[ftp://surfer.nmr.mgh.harvard.edu/pub/data/Yeo_CerebCortex2015_Brainmap.zip|Excel sheets of Pr(component | task) and Pr(activation | component) in FreeSurfer and MNI152 space]]. These components will be included in some future FreeSurfer release. Note however, the zip file linked here corresponds exactly to that from the reference, while components in the developmental and official release versions of FreeSurfer are subjected to modifications/improvements. |
| Line 23: | Line 23: |
| See README in unzipped folder | See README in unzipped folder. |
| Line 25: | Line 25: |
| == Parcellations in Nonlinear MNI152 Volume Space == ||'''7 Network Tight Mask''' ||'''7 Network Liberal Mask'''||'''17 Network Tight Mask''' ||'''17 Network Liberal Mask'''|| || {{attachment:Yeo2011_JNeurophysiol_7networks_TightMask_axial9.png}} || {{attachment:Yeo2011_JNeurophysiol_7networks_LiberalMask_axial9.png}}|| {{attachment:Yeo2011_JNeurophysiol_17networks_TightMask_axial9.png}} || {{attachment:Yeo2011_JNeurophysiol_17networks_LiberalMask_axial9.png}}|| |
Important Note: Because some image viewers do not handle these range of values very well, we have multiplied the values in the *PrActGivenComp* files by 1e5. Therefore in the resulting volumes, value of 1 corresponds to probability of 1e-5. |
| Line 29: | Line 27: |
| ==== Downloads ==== [[ftp://surfer.nmr.mgh.harvard.edu/pub/data/Yeo_JNeurophysiol11_MNI152.zip|Resting State Cortical Parcellation in nonlinear MNI152 space]]. We are working to put these subjects in FreeSurfer. Note however, the zip file linked here corresponds exactly to that from the reference, while functional parcellations in the developmental and official release versions of FreeSurfer are subjected to modifications/improvements. |
== Related downloads == [[https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation_Yeo2011|Resting State Cerebral Cortex Parcellations]] |
| Line 32: | Line 30: |
| ==== Information about Downloads ==== 1. `FSL_MNI152_FreeSurferConformed_1mm.nii.gz` is the FSL MNI152 1mm template interpolated and intensity normalized into a 256 x 256 x 256 1mm-isotropic volume (obtained by putting the FSL MNI152 1mm template through recon-all using FreeSurfer 4.5.0) |
[[http://www.youtube.com/user/YeoKrienen|Movies of Cortical Seed-based Resting State Functional Connectivity]] |
| Line 35: | Line 32: |
| 2. `Yeo2011_7Networks_MNI152_FreeSurferConformed1mm.nii.gz` is a volume consisting of 7 cortical networks projected into MNI152 space. The cortical ribbon is defined by putting the FSL MNI152 1mm template through recon-all using FreeSurfer 4.5.0. The slices of this volume is shown in Yeo et al., 2011. | [[https://surfer.nmr.mgh.harvard.edu/fswiki/CerebellumParcellation_Buckner2011|Resting State Cerebellum Parcellation]] |
| Line 37: | Line 34: |
| 3. `Yeo2011_7Networks_MNI152_FreeSurferConformed1mm.nii.gz` is a volume consisting of 7 cortical networks projected into MNI152 space. The cortical ribbon is defined in a more liberal fashion than in (2). More specifically, the cortical mask is obtained by nonlinear warping 1000 subjects (from Yeo et al. 2011, Buckner et al. 2011) into MNI152 space via the FreeSurfer recon-all pipeline. An initial mask is first obtained where a vox l is decided to be a cortical voxel if the cortex of more than 150 subjects (out of 1000 subjects) were mapped to the voxel or if the voxel is labeled as part of the cortical ribbon from (2). This cortical mask is th n smoothed, and small holes and islands in the masks are removed by simple morphological operations. 4. `Yeo2011_7Networks_ColorLUT.txt` is a FreeSurfer readable text file specifying how the 7 networks are named, numbered and colored in Yeo et al. 2011: || Index || Network Name || R || G || B || || || 0 || NONE || 0 || 0 || 0 || 0 || || 1 || 7Networks_1 || 120 || 18 || 134 || 0 || || 2 || 7Networks_2 || 70 || 130 || 180 || 0 || || 3 || 7Networks_3 || 0 || 118 || 14 || 0 || || 4 || 7Networks_4 || 196 || 58 || 250 || 0 || || 5 || 7Networks_5 || 220 || 248 || 164 || 0 || || 6 || 7Networks_6 || 230 || 148 || 34 || 0 || || 7 || 7Networks_7 || 205 || 62 || 78 || 0 || In particular the networks are numbered from 7Networks_1 to 7Networks_7. The first column of the text file specifies the value of voxels in the nifty values corresponding to the particular network. The second column f the text file specifies the named of the networks. For example, from the text file, voxels whose values = 3 corresponds to the network 7Networks_3.Columns 3 to 5 corresponds to the R, G, B values (ranges from 0 to 55) of the networks. Last column is all zeros (FreeSurfer's default). 5. `Yeo2011_17Networks_MNI152_FreeSurferConformed1mm.nii.gz` is a volume consisting of 17 cortical networks projected into MNI152 space. The cortical ribbon is defined in the same fashion as (2), i.e., by putting the F L MNI152 1mm template through recon-all using FreeSurfer 4.5.0. 6. `Yeo2011_17Networks_MNI152_FreeSurferConformed1mm.nii.gz` is a volume consisting of 17 cortical networks projected into MNI152 space. The cortical ribbon is defined in a more liberal fashion than in (2) and in the s me way as (3). 7. `Yeo2011_17Networks_ColorLUT.txt` is a FreeSurfer readable text file specifying how the 17 networks are named, numbered and colored in Yeo et al. 2011: || Index || Network Name || R || G || B || || || 0 || NONE || 0 || 0 || 0 || 0 || || 1 || 17Networks_1 || 120 || 18 || 134 || 0 || || 2 || 17Networks_2 || 255 || 0 || 0 || 0 || || 3 || 17Networks_3 || 70 || 130 || 180 || 0 || || 4 || 17Networks_4 || 42 || 204 || 164 || 0 || || 5 || 17Networks_5 || 74 || 155 || 60 || 0 || || 6 || 17Networks_6 || 0 || 118 || 14 || 0 || || 7 || 17Networks_7 || 196 || 58 || 250 || 0 || || 8 || 17Networks_8 || 255 || 152 || 213 || 0 || || 9 || 17Networks_9 || 200 || 248 || 164 || 0 || || 10 || 17Networks_10 || 122 || 135 || 50 || 0 || || 11 || 17Networks_11 || 119 || 140 || 176 || 0 || || 12 || 17Networks_12 || 230 || 148 || 34 || 0 || || 13 || 17Networks_13 || 135 || 50 || 74 || 0 || || 14 || 17Networks_14 || 12 || 48 || 255 || 0 || || 15 || 17Networks_15 || 0 || 0 || 130 || 0 || || 16 || 17Networks_16 || 255 || 255 || 0 || 0 || || 17 || 17Networks_17 || 205 || 62 || 78 || 0 || ==== Example Usage ==== See README in unzipped folder == Other downloads == [[http://sumsdb.wustl.edu:8081/sums/directory.do?id=8286317|Cerebral Surface Parcellations in Caret Space]] [[http://www.youtube.com/user/YeoKrienen|Movies of Cortical Seed-based Functional Connectivity]] [[http://surfer.nmr.mgh.harvard.edu/fswiki/CerebellumParcellation_Buckner2011|Cerebellar Parcellation in MNI Space]] [[http://www.youtube.com/BucknerKrienen|Movies of Cerebellar Seed-based Functional Connectivity]] [[http://surfer.nmr.mgh.harvard.edu/fswiki/StriatumParcellation_Choi2012| Striatum Parcellation in MNI Space]] [[http://people.csail.mit.edu/danial/Site/Code.html| Code for the von Mises-Fisher Mixture Model Clustering]] |
[[https://surfer.nmr.mgh.harvard.edu/fswiki/StriatumParcellation_Choi2012 | Resting State Striatum Parcellation]] |
Nested Cognitive Ontology Estimated From 10,449 Experiments
We applied a hierarchical Bayesian model (figure on right) on 10,449 Brainmap experiments across 83 behavioral tasks. By formalizing the notion that performing a task engages multiple cognitive components, each supported by overlapping brain regions, we identified cognitive components that are shared across tasks. Components divided into subcomponents as the number of estimated components increased, revealing a nested ontology. The components enabled the derivation of quantitative maps of functional specialization, revealing complex zones of frontal and parietal regions ranging from being highly specialized to highly flexible. The network organization of the specialized and flexible regions was explored with an independent resting-state fMRI dataset (N=1000). Cortical regions specialized for the same components were strongly coupled, suggesting that components function as partially isolated networks. Functionally flexible regions participate in multiple components to different degrees. This heterogeneous selectivity was strongly predicted by the connectivity between flexible and specialized regions. Functionally flexible regions might support the core of the brain’s information processing capacity, binding or integrating the processing power of segregated, specialized brain networks. The estimated components in FreeSurfer fsaverage space and MNI152 space, as well as specificity and flexibility estimates are available for download. |
|
References
Interactive 12-Component Cognitive Ontology
Interactive 12-component Ontology can be accessed here! We found the interactive version of the static ontology (below) extremely useful.
Downloads
[[ftp://surfer.nmr.mgh.harvard.edu/pub/data/Yeo_CerebCortex2015_Brainmap.zip|Excel sheets of Pr(component | task) and Pr(activation | component) in FreeSurfer and MNI152 space]]. These components will be included in some future FreeSurfer release. Note however, the zip file linked here corresponds exactly to that from the reference, while components in the developmental and official release versions of FreeSurfer are subjected to modifications/improvements.
Example Usage
See README in unzipped folder.
Important Note: Because some image viewers do not handle these range of values very well, we have multiplied the values in the *PrActGivenComp* files by 1e5. Therefore in the resulting volumes, value of 1 corresponds to probability of 1e-5.
Related downloads
Resting State Cerebral Cortex Parcellations
Movies of Cortical Seed-based Resting State Functional Connectivity