| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
|
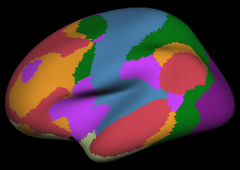
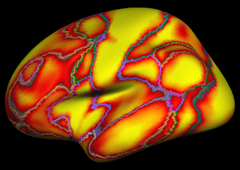
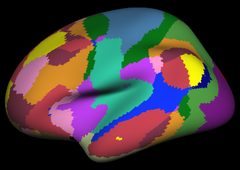
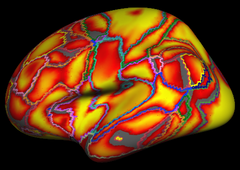
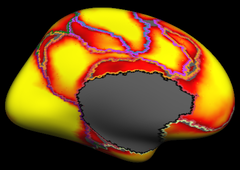
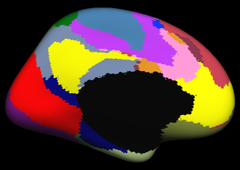
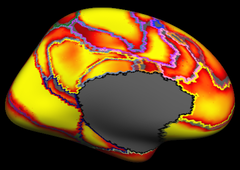
Resting state fMRI data from 1000 subjects were registered using surface-based alignment. A clustering approach was employed to identify and replicate networks of functionally coupled regions across the cerebral cortex. The results revealed local networks confined to sensory and motor cortices as well as distributed networks of association regions. Within the sensory and motor cortices, functional connectivity followed topographic representations across adjacent areas. In association cortex, the connectivity patterns often showed abrupt transitions between network boundaries. |
Resting State Cortical Parcellation
Resting state fMRI data from 1000 subjects were registered using surface-based alignment. A clustering approach was employed to identify and replicate networks of functionally coupled regions across the cerebral cortex. The results revealed local networks confined to sensory and motor cortices as well as distributed networks of association regions. Within the sensory and motor cortices, functional connectivity followed topographic representations across adjacent areas. In association cortex, the connectivity patterns often showed abrupt transitions between network boundaries.
7 Network Estimate |
7 Network Confidence |
17 Network Estimate |
17 Network Confidence |
|
|
|
|
|
|
|
|
Downloads
Resting State Cortical Parcellation in fsaverage, fsaverage6 and fsaverage5 Space. These subjects are also currently in the developmental version of FreeSurfer which can be assessed from the Martinos center network (/autofs/cluster/freesurfer/centos4.0_x86_64/dev/subjects/). These subjects will be officially included in some future FreeSurfer release.
Movies of Seed-based Functional Connectivity
Information about Downloads
There are three folders in "Yeo_JNeurophysiol11_FreeSurfer.zip", corresponding to the "fsaverage", "fsaverage5" and "fsaverage6" surface space. "fsaverage" contains the high resolution version of the parcellation, while "fsaverage6" and "fsaverage5" contain lower resolution versions of the parcellation. The parcellations were computed in "fsaverage5" space and upsampled to "fsaverage6" and "fsaverage".
The structure of each folder follows that of a preprocessed freesurfer subject. In particular, "fsaverage/label/", "fsaverage5/label/", "fsaverage6/label/" contain all the parcellation and confidence files. For example, "fsaverage/label/rh.Yeo2011_7Networks_N1000.annot" is the 7-network parcellation for 1000 subjects on the right hemisphere and "fsaverage/label/lh.Yeo2011_17NetworksConfidence_N1000.mgz" is the confidence map for the 17-network parcellation for 1000 subjects on the left hemisphere.