|
Size: 779
Comment:
|
Size: 2212
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| see https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation_Yeo2011 for example | |
| Line 4: | Line 3: |
| intro here, | {{attachment:flateconlyinflated2014.jpg}} |
| Line 6: | Line 5: |
| ''Author: Jean Augustinack'' | |
| Line 7: | Line 7: |
| The entorhinal cortex is located on the crown of the anterior parahippocampal gyrus. Brodmann assigned the number 28 to entorhinal cortex (Brodmann’s area 28). Entorhinal cortex displays a unique cytoarchitecture with large neurons in layer II that form clusters. The entorhinal cortex is the gateway to the hippocampus. In Alzheimer’s disease (AD), neurons in the entorhinal cortex, particularly those in layer II, begin to die and some leave behind a pathological marker – the neurofibrillary tangle. Neurofibrillary tangles and significant cell death characterize the neuronal loss in AD where neurofibrillary tangles and subsequent atrophy have been correlated with dementia. | |
| Line 8: | Line 9: |
| you have three options for using these labels: | Please note that there two entorhinal labels exist in FreeSurfer 1) the EC label included in the aparc atlas and 2) the cytoarchitecturally-defined entorhinal label (i.e., the ex vivo one). |
| Line 10: | Line 11: |
| labels in freesurfer surface space are in subjects dir subj/label/?h.perirhinal.label | Reference for Entorhinal cortex, Cytoarchitecturally-defined labels: |
| Line 12: | Line 13: |
| instructions on how to view in freeview | Fischl B, et al. Predicting the location of entorhinal cortex from MRI. Neuroimage. 2009 Aug 1;47(1):8-17. |
| Line 14: | Line 15: |
| where to get stats on this label (../stats/$hemi.BA.stats) | == Where to find the entorhinal (cytoarchitectural-defined) labels in FreeSurfer == |
| Line 16: | Line 17: |
| already in freesurfer 5.3 | Right and left labels for entorhinal are available in freesurfer 5.3 and 6.0+. |
| Line 18: | Line 19: |
| The labels are in freesurfer surface space and located in subjects dir: {{{ subj/label/?h.entorhinal_exvivo.label }}} |
|
| Line 19: | Line 24: |
| They are generated by default by: {{{ recon-all -all }}} but can be regenerated with: {{{ recon-all -ba-labels }}} |
|
| Line 20: | Line 33: |
| label in volume space | In freeview, after loading a surface, the label can be viewed either by loading the Annotation file: {{{ subj/label/?h.BA_exvivo.annot }}} or by loading the Label file: {{{ subj/label/?h.entorhinal_exvivo.label }}} (which allows selection of 'Heat Scale' viewing mode to display the probability mapping), or by loading the Label file: {{{ subj/label/?h.entorhinal_exvivo.thresh.label }}} which is a thresholded version, more suitable as an ROI. |
| Line 22: | Line 47: |
| mapping surface label to subjects volume space | Stats on this label are found in: {{{ subj/stats/?h.BA_exvivo.stats }}} |
| Line 24: | Line 52: |
| will be available in v6 or if you have freesurfer, run this to map from surface space to volume space and then this is how you view it. label in fsaverage volume space [[attachment:perirhinal_fsaverage.nii]] label in mni152 volume space [[attachment:perirhinal_mni152.nii]] |
See also: * [[BrodmannAreaMaps]] * [[Perirhinal]] * [[ExVivoEcPcLabelsFsaverage]] |
Entorhinal Cortex
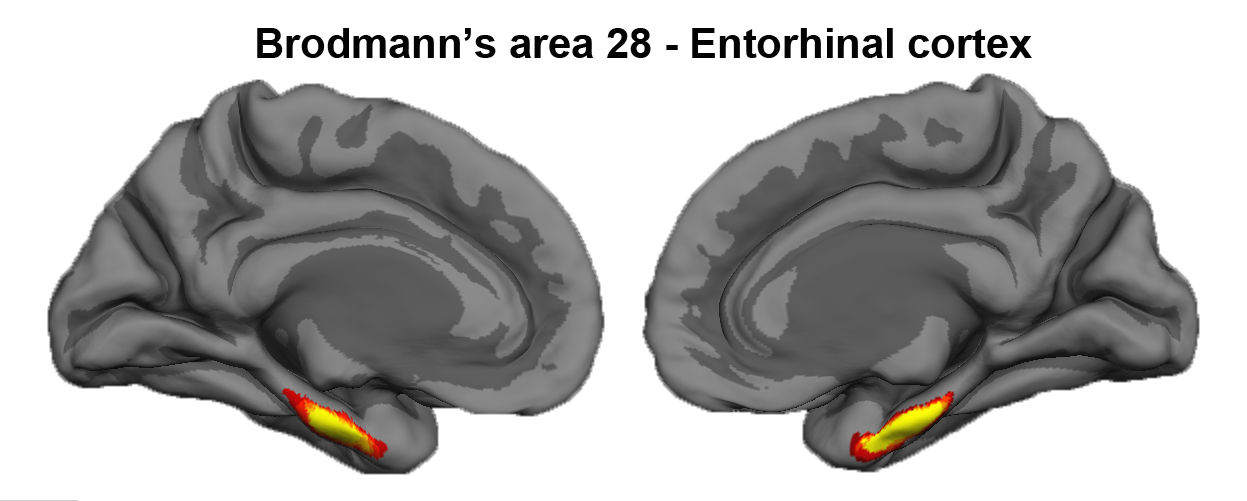
Author: Jean Augustinack
The entorhinal cortex is located on the crown of the anterior parahippocampal gyrus. Brodmann assigned the number 28 to entorhinal cortex (Brodmann’s area 28). Entorhinal cortex displays a unique cytoarchitecture with large neurons in layer II that form clusters. The entorhinal cortex is the gateway to the hippocampus. In Alzheimer’s disease (AD), neurons in the entorhinal cortex, particularly those in layer II, begin to die and some leave behind a pathological marker – the neurofibrillary tangle. Neurofibrillary tangles and significant cell death characterize the neuronal loss in AD where neurofibrillary tangles and subsequent atrophy have been correlated with dementia.
Please note that there two entorhinal labels exist in FreeSurfer 1) the EC label included in the aparc atlas and 2) the cytoarchitecturally-defined entorhinal label (i.e., the ex vivo one).
Reference for Entorhinal cortex, Cytoarchitecturally-defined labels:
Fischl B, et al. Predicting the location of entorhinal cortex from MRI. Neuroimage. 2009 Aug 1;47(1):8-17.
Where to find the entorhinal (cytoarchitectural-defined) labels in FreeSurfer
Right and left labels for entorhinal are available in freesurfer 5.3 and 6.0+.
The labels are in freesurfer surface space and located in subjects dir:
subj/label/?h.entorhinal_exvivo.label
They are generated by default by:
recon-all -all
but can be regenerated with:
recon-all -ba-labels
In freeview, after loading a surface, the label can be viewed either by loading the Annotation file:
subj/label/?h.BA_exvivo.annot
or by loading the Label file:
subj/label/?h.entorhinal_exvivo.label
(which allows selection of 'Heat Scale' viewing mode to display the probability mapping), or by loading the Label file:
subj/label/?h.entorhinal_exvivo.thresh.label
which is a thresholded version, more suitable as an ROI.
Stats on this label are found in:
subj/stats/?h.BA_exvivo.stats
See also:
