|
Size: 1789
Comment:
|
← Revision 9 as of 2008-04-29 11:46:03 ⇥
Size: 2183
Comment: converted to 1.6 markup
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| This process uses the command mri_fill to create a binarized volume. The input is the mri/wm volume and the output is the mri/filled volume. First, the cutting planes are determined to seperate the hemispheres from each other and the pons. Then a seed point is located and used to fill each hemisphere. The rh will be filled with a value of 255, and the lh with a value of 127. | This process uses the command [[mri_fill]] to create a binarized volume. The input is the mri/wm volume and the output is the mri/filled volume. First, the cutting planes are determined to separate the hemispheres from each other and the pons. Then a seed point is located and used to fill each hemisphere. The rh will be filled with a value of 255, and the lh with a value of 127. |
| Line 3: | Line 3: |
| Occasionnally, manual intervention is required to define the cutting planes and seed points. To do so, follow these steps: | Occasionally, manual intervention is required to define the cutting planes and seed points. To do so, follow these steps: |
| Line 5: | Line 5: |
| 1.Open the subject's wm volume up in tkmedit. | 1. Open the subject's (subj) wm volume up in [[tkmedit]]. |
| Line 7: | Line 7: |
| '''tkmedit subject_name wm''' | [[tkmedit]] subj wm |
| Line 9: | Line 9: |
| 2.View the MNI talairach coordinates. | 1. View the MniTalairach coordinates: '''View->Information->MNI Coordinates''' |
| Line 11: | Line 11: |
| '''View->Information->MNI Coordinates''' | 1. Select the Corpus Callosum point, on the mid-posterior portion of it. Example: |
| Line 13: | Line 13: |
| 3.Select the Corpus Callosum point, on the mid-posterior portion of it. Example: | Corpus Callosum |
| Line 15: | Line 15: |
| attachment:CCcor.jpg attachment:CChor.jpg attachment:CCsag.jpg | {{attachment:CCcor.jpg}} {{attachment:CChor.jpg}} {{attachment:CCsag.jpg}} |
| Line 17: | Line 17: |
| 4.Copy the MNI coordinates to the command line for either '''recon-all -cc-xyz ....''' or in '''mri_fill -C ....''' | 1. Copy the MNI coordinates to the command line for either |
| Line 19: | Line 19: |
| 5.Do this for all of the other cutting plane points and seed points. Examples follow: | [[recon-all]] -cc-xyz .... |
| Line 21: | Line 21: |
| Pons | or in |
| Line 23: | Line 23: |
| attachment:Pcor.jpg attachment:Phor.jpg attachment:Psag.jpg | [[mri_fill]] -C .... |
| Line 25: | Line 25: |
| RH seed | Alternatively seed point can be given in voxel coordinates under the command line [[mri_fill]] : -CV <x> <y> <z> - the voxel coords of the seed for the corpus callosum -PV <x> <y> <z> - the voxel coords of the seed for the pons |
| Line 27: | Line 31: |
| attachment:RHcor.jpg attachment:RHhor.jpg attachment:RHsag.jpg | 1. Do this for all of the other cutting plane points and seed points. Examples follow: |
| Line 29: | Line 33: |
| LH seed | Pons |
| Line 31: | Line 35: |
| attachment:LHcor.jpg attachment:LHhor.jpg attachment:LHsag.jpg | {{attachment:Pcor.jpg}} {{attachment:Phor.jpg}} {{attachment:Psag.jpg}} |
| Line 33: | Line 37: |
| 6.Not all points need to be defined, any range from none to all four may be defined. in mri_fill: | RH seed |
| Line 35: | Line 39: |
| {{{mri_fill -C MNIcoords -P MNIcoords -rh MNIcoords -lh MNIcoords $SUBJECTS_DIR/subj/mri/wm $SUBJECTS_DIR/subj/mri/filled;inflate_subject-rh subj;inflate_subject-lh subj}}} | {{attachment:RHcor.jpg}} {{attachment:RHhor.jpg}} {{attachment:RHsag.jpg}} |
| Line 37: | Line 41: |
| LH seed | |
| Line 38: | Line 43: |
| or in the equivalent in recon-all: | {{attachment:LHcor.jpg}} {{attachment:LHhor.jpg}} {{attachment:LHsag.jpg}} |
| Line 40: | Line 45: |
| {{{recon-all -stage2 -cc-xyz MNIcoords -pons-xyz MNIcoords -rh-xyz MNIcoords -lh-xyz MNIcoords -subjid subj}}} | 1. Not all points need to be defined, any range from none to all four may be defined. To inflate: <<BR>> <<BR>> [[mri_fill]] -C MNIcoords -P MNIcoords -rh MNIcoords -lh MNIcoords $SUBJECTS_DIR/subj/mri/wm $SUBJECTS_DIR/subj/mri/filled;inflate_subject-rh subj;inflate_subject-lh subj <<BR>> <<BR>> |
| Line 42: | Line 50: |
| '''NOTE: these command line(s) need to be run EVERYTIME the subject's surfaces are reinflated''' | or, the equivalent in recon-all: <<BR>><<BR>> [[recon-all]] -stage2 -cc-xyz MNIcoords -pons-xyz MNIcoords -rh-xyz MNIcoords -lh-xyz MNIcoords -subjid subj <<BR>><<BR>> |
| Line 44: | Line 55: |
| '''NOTE: These command line(s) need to be run EVERYTIME the subject's surfaces are reinflated''' |
This process uses the command mri_fill to create a binarized volume. The input is the mri/wm volume and the output is the mri/filled volume. First, the cutting planes are determined to separate the hemispheres from each other and the pons. Then a seed point is located and used to fill each hemisphere. The rh will be filled with a value of 255, and the lh with a value of 127.
Occasionally, manual intervention is required to define the cutting planes and seed points. To do so, follow these steps:
Open the subject's (subj) wm volume up in tkmedit.
tkmedit subj wm
View the MniTalairach coordinates: View->Information->MNI Coordinates
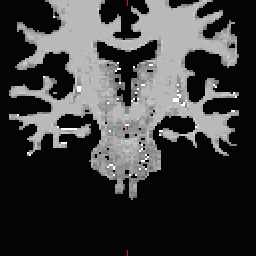
- Select the Corpus Callosum point, on the mid-posterior portion of it. Example: Corpus Callosum
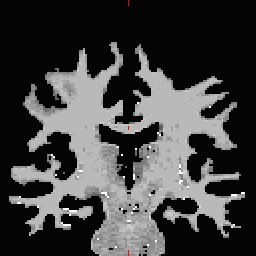
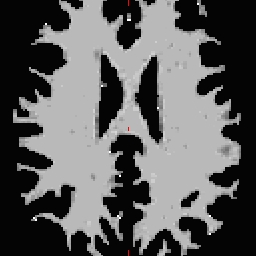
- Copy the MNI coordinates to the command line for either
recon-all -cc-xyz .... or in
mri_fill -C ....
Alternatively seed point can be given in voxel coordinates under the command line mri_fill :
-CV <x> <y> <z> - the voxel coords of the seed for the corpus callosum
-PV <x> <y> <z> - the voxel coords of the seed for the pons
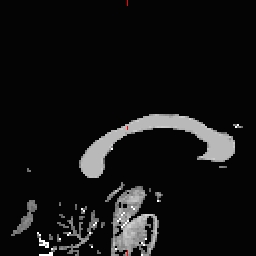
- Do this for all of the other cutting plane points and seed points. Examples follow: Pons
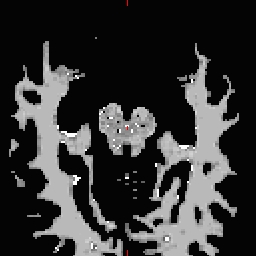
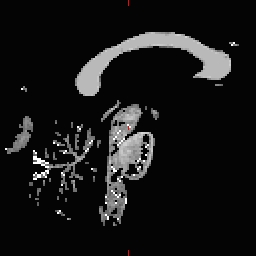 RH seed
RH seed 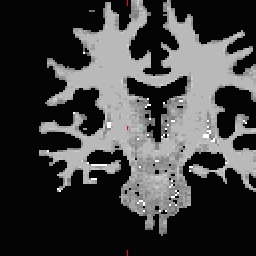
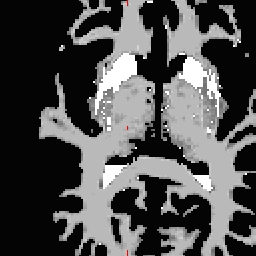
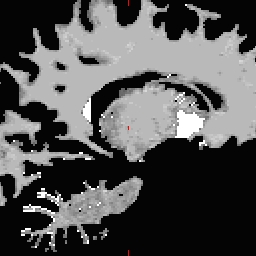 LH seed
LH seed 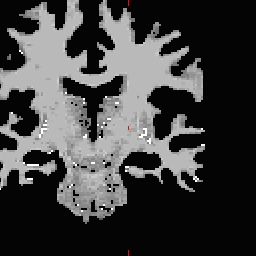
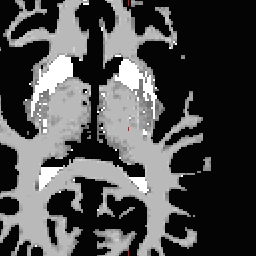
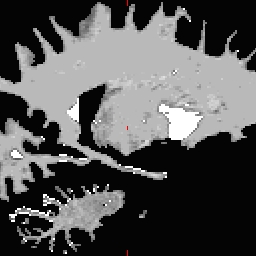
- Not all points need to be defined, any range from none to all four may be defined. To inflate:
mri_fill -C MNIcoords -P MNIcoords -rh MNIcoords -lh MNIcoords $SUBJECTS_DIR/subj/mri/wm $SUBJECTS_DIR/subj/mri/filled;inflate_subject-rh subj;inflate_subject-lh subj
or, the equivalent in recon-all:
recon-all -stage2 -cc-xyz MNIcoords -pons-xyz MNIcoords -rh-xyz MNIcoords -lh-xyz MNIcoords -subjid subj
NOTE: These command line(s) need to be run EVERYTIME the subject's surfaces are reinflated
