|
Size: 16080
Comment:
|
Size: 16148
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 270: | Line 270: |
| {{attachment:path.waypts.jpg}} {{attachment:path.waypts.p.jpg}} |
Tract statistics
Remember...
For each new terminal that you open, you must do:
setenv SUBJECTS_DIR $TUTORIAL_DATA/diffusion_recons cd $TUTORIAL_DATA/diffusion_tutorial
This section of the tutorial will teach you how to extract statistics on anisotropy and diffusivity measures for the white-matter pathways reconstructed by TRACULA. There are two types of statistics files that are created for each white-matter pathway:
- pathstats.overall.txt - This file contains measures averaged over the whole pathway
- pathstats.byvoxel.txt - This file contains measures as a function of position along the trajectory of the pathway.
For example, the statistics files for the left inferior longitudinal fasciculus (ILF) of subject elmo.2012, showing measures for the entire left ILF or along the left ILF, are, respectively:
$TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt
Anisotropy and diffusivity averaged over an entire pathway
You can view an example of the overall pathway stats by doing:
gedit $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt &
This file will look like this:
# Title Pathway Statistics # # generating_program /usr/local/freesurfer/dev/bin/dmri_pathstats # cvs_version # cmdline /usr/local/freesurfer/dev/bin/dmri_pathstats --intrc /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr --dtbase /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dmri/dtifit --path lh.ilf --subj elmo.2012 --out /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --outvox /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt # sysname Linux # hostname compute-0-29 # machine x86_64 # user ayendiki # anatomy_type pathway # # subjectname elmo.2012 # pathwayname lh.ilf # Count 1500 Volume 167 Len_Min 29 Len_Max 64 Len_Avg 45.008 Len_Center 37 AD_Avg 0.00131194 AD_Avg_Weight 0.00139891 AD_Avg_Center 0.001268 RD_Avg 0.000686486 RD_Avg_Weight 0.000770628 RD_Avg_Center 0.000635293 MD_Avg 0.00089497 MD_Avg_Weight 0.000980054 MD_Avg_Center 0.000846196 FA_Avg 0.414059 FA_Avg_Weight 0.392266 FA_Avg_Center 0.408256
This text file contains various diffusion measures, averaged over the entire white-matter tract. The measures include:
- Number of sample paths in the WM tract
- Tract volume (in voxels)
- Maximum, minimum and average length of sample paths
- Length of the highest-probability (a.k.a. maximum a posteriori) path
- Axial diffusivity (average over the entire support of the path distribution, weighted average over the entire support of the path distribution, and average over highest-probability path only)
- Radial diffusivity (as above)
- Mean diffusivity (as above)
- Fractional anisotropy (as above)
Converting pathstats.overall.txt files to a table for group analyses
Measures can be extracted from these files to be analyzed further, e.g., for tract-based group analysis. Specifically, the text files can be converted into a table using the command tractstats2table and then used for doing GLM analyses with mri_glmfit or any other statistical software (SPSS, Excel, Statview etc.)
To extract all diffusion measures for the left ILF from subject elmo.2012 into a table, do the following:
tractstats2table --inputs $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --overall --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table
The argument to the --inputs option specifies the input stats file.
The --overall option tells tractstats2table to expect the overall path stats file (stats averaged over the entire left ILF), as opposed to the stats along the trajectory of the tract (which would be specified with the --byvoxel option instead).
The argument to the --tablefile option specifies the output file.
Take a look at the resulting table file:
gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table &
For a group analysis, tractstats2table can be used to combine the diffusion measures in pathstats.overall.txt from multiple subjects into a single table, which can then be used for analysis with mri_glmfit or any other statistical software.
To do this, you will have to create a text file that lists the full path to every subject's pathstats.overall.txt file. An example of such a list for all 3 tutorial subjects is shown below:
$TUTORIAL_DATA/diffusion_tutorial/elmo.2005/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2008/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt
Create a text file that contains the list above and name it as follows:
gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list &
The following command will use this list to create a table with the diffusion measures from all the files listed above:
tractstats2table --load-pathstats-from-file $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list --overall --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table
The argument to the --load-pathstats-from-file option specifies the text file that contains the list of all the statistics files that will be loaded.
Take a look at the resulting table file:
gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table &
Instead of extracting all measures included in the tract statistics files, we may be interested only in a few specific measures. For example, to extract only the average FA for each subject, do the following:
tractstats2table --load-pathstats-from-file $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list --overall --only-measures FA_Avg --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.FA_Avg.table
The argument to the --only-measures option specifies which measure we want to extract from the statistics files. Instead of FA_Avg, this could be the name of any of the measures included in pathstats.overall.txt.
You can look at these tables in OpenOffice (or any other spreadsheet program). For example, to open the file lh.ilf.All.table in OpenOffice, do the following:
oocalc $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table
From here, you could use these stats tables to run a group analysis on the tracts.
Anisotropy and diffusivity along the trajectory of a pathway
Now take a look at the stats as a function of position along the trajectory of the same tract:
gedit $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt &
It will look like this:
# Title Pathway Statistics # # generating_program /usr/local/freesurfer/dev/bin/dmri_pathstats # cvs_version # cmdline /usr/local/freesurfer/dev/bin/dmri_pathstats --intrc /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr --dtbase /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dmri/dtifit --path lh.ilf --subj elmo.2012 --out /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --outvox /space/freesurfer/subjects/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt # sysname Linux # hostname compute-0-29 # machine x86_64 # user ayendiki # anatomy_type pathway # # subjectname elmo.2012 # pathwayname lh.ilf # # pathway start x y z AD RD MD FA AD_Avg RD_Avg MD_Avg FA_Avg 86 71 16 0.00124639 0.000810355 0.000955702 0.296343 0.00124166 0.000683586 0.000869599 0.375502 86 72 15 0.000758499 0.000690337 0.000713058 0.0665528 0.00120352 0.000674202 0.00085063 0.365702 85 72 14 0.000819579 0.000601256 0.00067403 0.196614 0.00111348 0.000618912 0.00078378 0.353662 84 72 13 0.000970979 0.00067094 0.000770953 0.227346 0.00115739 0.000679319 0.000838683 0.329086 84 71 13 0.0010519 0.000845654 0.000914402 0.174291 0.00117946 0.000675255 0.000843332 0.342993 84 70 13 0.00122524 0.00075747 0.000913395 0.295734 0.00127004 0.000700568 0.00089039 0.368428 84 69 13 0.0013762 0.000607907 0.000864005 0.475513 0.00130121 0.000666051 0.000877759 0.405086 83 68 14 0.00117165 0.000572571 0.000772264 0.433612 0.00127088 0.000631783 0.000844803 0.427719 83 67 14 0.00122987 0.000573616 0.000792367 0.452554 0.00123866 0.000625767 0.000830053 0.422219 83 66 14 0.00120132 0.000599483 0.000800095 0.411401 0.00118774 0.00062832 0.000814796 0.408966 83 65 14 0.00130532 0.000741545 0.000929471 0.341101 0.00117765 0.000649599 0.000825623 0.38951 83 64 15 0.00113069 0.000744614 0.000873306 0.303024 0.00124994 0.000716112 0.000894054 0.35574 83 63 15 0.00145392 0.000843014 0.00104665 0.338389 0.00160184 0.000956015 0.00117128 0.342898 83 62 15 0.00150758 0.000704525 0.000972208 0.454268 0.00184354 0.00110231 0.0013494 0.361096 82 61 15 0.00168556 0.000719335 0.00104141 0.500387 0.00187725 0.00105482 0.00132898 0.413339 82 60 16 0.00149165 0.000655993 0.000934546 0.482961 0.00174103 0.000915708 0.00119082 0.459582 82 59 16 0.00178469 0.000676534 0.00104592 0.547817 0.00181009 0.000942766 0.00123188 0.47797 82 58 16 0.00187731 0.000555021 0.000995783 0.650825 0.00176598 0.000879462 0.00117498 0.482668 82 57 17 0.00136583 0.000342585 0.000683666 0.706805 0.00183719 0.00101521 0.00128919 0.444575 82 56 17 0.00149454 0.000361014 0.000738857 0.718399 0.00172875 0.000879471 0.00116258 0.466697 82 55 17 0.00164246 0.000404404 0.000817091 0.714028 0.00143606 0.000654734 0.000915172 0.485324 81 54 17 0.00110069 0.000505138 0.000703656 0.462108 0.00123971 0.000546461 0.000777551 0.491684 81 53 18 0.00149581 0.000503958 0.000834577 0.601599 0.00138836 0.000547033 0.000827479 0.539068 81 52 18 0.00130913 0.000454745 0.00073954 0.597531 0.00133457 0.000539894 0.000804789 0.533393 81 51 18 0.00131507 0.000631822 0.000859572 0.448947 0.00131177 0.000606845 0.000841826 0.469117 81 50 18 0.00135444 0.000747886 0.000950069 0.362522 0.00129975 0.000659834 0.000873148 0.416883 81 49 18 0.00120955 0.000643043 0.000831879 0.404422 0.00126551 0.00064144 0.000849466 0.418424 80 48 19 0.00140649 0.000619495 0.000881827 0.476057 0.00134196 0.000607405 0.000852266 0.47251 80 47 19 0.00124703 0.00054436 0.000778584 0.493392 0.00129913 0.000595745 0.000830194 0.470203 80 46 19 0.00123713 0.000562425 0.000787325 0.473597 0.00131166 0.000608192 0.000842689 0.465411 80 45 19 0.00131702 0.000638351 0.000864576 0.436198 0.00131184 0.000638683 0.000863079 0.443722 79 44 20 0.000961184 0.000655564 0.000757438 0.251941 0.00120554 0.000691043 0.000862543 0.349984 79 43 20 0.00104516 0.000760429 0.000855339 0.211843 0.00118236 0.000713172 0.000869568 0.323664 79 42 20 0.0010671 0.000695726 0.000819518 0.290868 0.00118518 0.000688767 0.000854257 0.347132 79 41 20 0.000951325 0.000643122 0.000745857 0.290616 0.00115072 0.000662836 0.000825473 0.352023 80 40 20 0.00111846 0.000771887 0.00088741 0.232294 0.00118544 0.000701789 0.000863008 0.328079 81 40 20 0.000989299 0.000649715 0.00076291 0.283576 0.00114976 0.000669422 0.000829546 0.343642 # pathway end
This text file contains various diffusion measures, one row for each position along the trajectory of the path. The first three entries in each row are the x, y, z coordinates in native diffusion space. The next four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at that position on the maximum a posteriori path. The last four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at the same position, averaged over all sampled paths.
Converting pathstats.byvoxel.txt files to a table for group analyses
The pathstats.byvoxel.txt files will generally not contain the same number of positions (rows) for each subject because the tracts are reconstructed in each subject's native diffusion space and not in a template space. Thus they are not ready for performing group analyses yet. To combine these files from multiple subjects, interpolating the anisotropy and diffusivity values at corresponding positions along the tract for all subjects, run the following:
trac-all -stat -c $TUTORIAL_DATA/diffusion_tutorial/dmrirc.tutorial
This will create a directory named stats under the main TRACULA output directory and save one table per tract per diffusion measure. In these tables, each row is a different position along the trajectory of the tract and each column is a different subject.
To examine the table of average FA along the left ILF, do the following:
gedit $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.FA_Avg.txt &
The contents of this file will look like this:
elmo.2005 elmo.2008 elmo.2012 NaN 0.335817 NaN 0.373068 0.35773 0.263802 0.37555 0.372835 0.280039 0.389334 0.373771 0.304207 0.403752 0.36399 0.340167 0.418984 0.363519 0.333422 0.433291 0.378936 0.356918 0.441835 0.397896 0.355258 0.432407 0.398355 0.356802 0.428443 0.399408 0.381642 0.428464 0.406425 0.410158 0.417515 0.415535 0.428911 ...
The first row tells you the subject name for the corresponding column of FA values. Note that for some data sets, there will be some NaN ("not a number") values in the beginning or end of the tract, because the tract has a slightly different length for each data set and the endings will not always correspond. For the purposes of a statistical analysis, you can ignore the endings, use only the data points that are not NaN, or pool together the data points from the first few or last few positions.
From here, you could use these stats tables to run a group analysis on the tracts.
Note: In addition to the above tables, the stats directory will also contain a log file for each pathway. You can examine the log file, which contains the full output of the trac-all -stat command, to see if any pathways were flagged as outliers. If this happens, there will be a line in the log file with "Found outlier path:" and the name of the subject. These are paths that were found to differ excessively from those of the other subjects. You can check this information to find out if the reconstruction of this tract failed for any subjects.
For example, you can examine the log file for the left ILF by doing:
gedit $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.log &
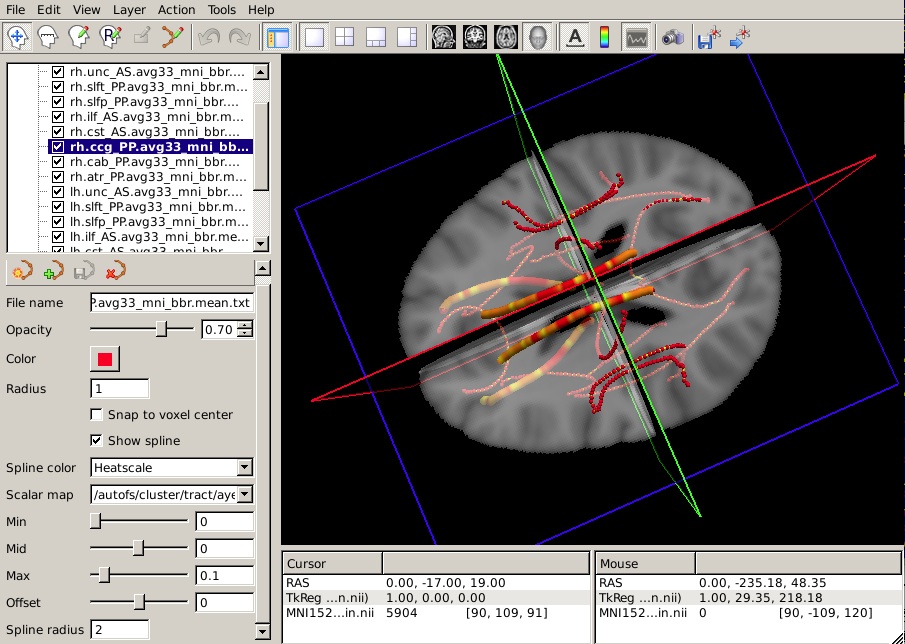
Visualizing results from statistical analyses along each pathway
The trac-all -stat command will also produce average paths in template space (MNI or CVS, depending on what was specified for the inter-subject registration method). These paths can be used to visualize the outputs of statistical analyses along each of the 18 tracts. Note that these average paths are only produced for visualization purposes and their existence does not imply that a voxel-based analysis was performed in template space in these coordinates. (Remember that TRACULA performs both the tractography and the extraction of anisotropy and diffusivity measures in the native diffusion space of each subject.)
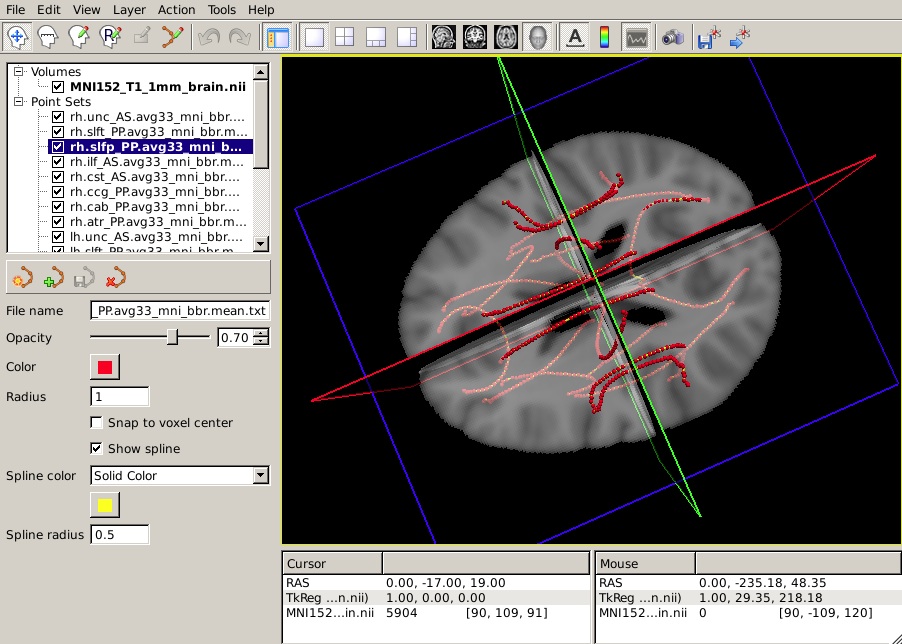
To view all 18 mean paths in the space of the MNI template, run the following:
freeview -v $FSLDIR/data/standard/MNI152_T1_1mm_brain.nii.gz \
-w $TUTORIAL_DATA/diffusion_tutorial/stats/*.path.mean.txt