|
Size: 6889
Comment:
|
Size: 21698
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| == Tract Statistics == This section of the tutorial will teach you to extract Statistical measures along the major White Matter tracts reconstructed by Tracula. There are two types of statistics files that are output for each White Matter Tract * path.overall.txt - This file gives Diffusion measures along the whole WM tract * path.byvoxel.txt - This files gives Diffusion measures for each voxel along the WM tract. To view the statistics files from Tracula for for the subject Diff001 (Left Cortico-Spinal Tract), type the following in the terminal |
## page was renamed from FsTutorial/TractStatistics [[https://surfer.nmr.mgh.harvard.edu/fswiki/Tutorials|Back to list of all tutorials]] | [[FsTutorial|Back to course page]] | [[FsTutorial/TraculaOutputs|Previous]] = Tract statistics = === Remember... === For '''each new''' terminal that you open, you must do: {{{ export SUBJECTS_DIR=$TUTORIAL_DATA/diffusion_recons cd $TUTORIAL_DATA/diffusion_tutorial }}} ------- This section of the tutorial will teach you how to extract statistics on anisotropy and diffusivity measures for the white-matter pathways reconstructed by TRACULA. There are two types of statistics files that are created for each white-matter pathway: * pathstats.overall.txt - This file contains measures averaged over the whole pathway * pathstats.byvoxel.txt - This file contains measures as a function of position along the trajectory of the pathway. For example, the statistics files for the left inferior longitudinal fasciculus (ILF) of subject elmo.2012, showing measures for the entire left ILF or along the left ILF, can be found below. '''Optional:''' use a text editor, such as {{{gedit}}}, or the command {{{open -e}}} on a Mac, to open these files. {{{ $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt }}} == Anisotropy and diffusivity averaged over an entire pathway == You can view an example of the overall pathway stats by doing: {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt & }}} This file will look like this: {{{ # Title Pathway Statistics # # generating_program /usr/local/freesurfer/stable6/bin/dmri_pathstats # cvs_version # cmdline /usr/local/freesurfer/stable6/bin/dmri_pathstats --intrc /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr --dtbase /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dmri/dtifit --path lh.ilf --subj elmo.2012 --out /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --outvox /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt # sysname Linux # hostname oribi # machine x86_64 # user eboyd2 # anatomy_type pathway # # subjectname elmo.2012 # pathwayname lh.ilf # Count 1500 Volume 629 Len_Min 36 Len_Max 71 Len_Avg 50.01 Len_Center 48 AD_Avg 0.00136398 AD_Avg_Weight 0.00137336 AD_Avg_Center 0.00137163 RD_Avg 0.000570444 RD_Avg_Weight 0.000568755 RD_Avg_Center 0.00056212 MD_Avg 0.000834957 MD_Avg_Weight 0.000836957 MD_Avg_Center 0.000831957 FA_Avg 0.505701 FA_Avg_Weight 0.510267 FA_Avg_Center 0.514496 }}} This text file contains various diffusion measures, averaged over the entire white-matter tract. The measures include: * Number of sample paths in the WM tract * Tract volume (in voxels) * Maximum, minimum and average length of sample paths * Length of the highest-probability (a.k.a. maximum a posteriori) path * Axial diffusivity (average over the entire support of the path distribution, weighted average over the entire support of the path distribution, and average over highest-probability path only) * Radial diffusivity (as above) * Mean diffusivity (as above) * Fractional anisotropy (as above) === Converting pathstats.overall.txt files to a table for group analyses === Measures can be extracted from these files to be analyzed further, e.g., for tract-based group analysis. Specifically, the text files can be converted into a table using the command {{{tractstats2table}}} and then used for doing GLM analyses with [[mri_glmfit]] or any other statistical software (SPSS, Excel, Statview etc.) To extract all diffusion measures for the left ILF from subject elmo.2012 into a table, do the following: {{{ tractstats2table --inputs $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --overall --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table }}} * The argument to the {{{--inputs}}} option specifies the input stats file. * The {{{--overall}}} option tells tractstats2table to expect the overall path stats file (stats averaged over the entire left ILF), as opposed to the stats along the trajectory of the tract (which would be specified with the {{{--byvoxel}}} option instead). * The argument to the {{{--tablefile}}} option specifies the output file. Take a look at the resulting table file: {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table & }}} For a group analysis, {{{tractstats2table}}} can be used to combine the diffusion measures in {{{pathstats.overall.txt}}} from multiple subjects into a single table, which can then be used for analysis with [[mri_glmfit]] or any other statistical software. To do this, you will have to create a text file that lists the full path to every subject's {{{pathstats.overall.txt}}} file. An example of such a list for all 3 tutorial subjects is shown below. {{{ $TUTORIAL_DATA/diffusion_tutorial/elmo.2005/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2008/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt }}} Create a text file using the command below. Copy and paste this list into it, and save the file. {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list & }}} The following command will use this list to create a table with the diffusion measures from all the files listed above: {{{ tractstats2table --load-pathstats-from-file $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list --overall --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table }}} * The argument to the {{{--load-pathstats-from-file}}} option specifies the text file that contains the list of all the statistics files that will be loaded. Take a look at the resulting table file: {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table & }}} Instead of extracting all measures included in the tract statistics files, we may be interested only in a few specific measures. For example, to extract only the average FA for each subject, do the following: {{{ tractstats2table --load-pathstats-from-file $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list --overall --only-measures FA_Avg --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.FA_Avg.table }}} * The argument to the {{{--only-measures}}} option specifies which measure we want to extract from the statistics files. Instead of FA_Avg, this could be the name of any of the measures included in {{{pathstats.overall.txt.}}} '''OPTIONAL:''' You can look at these tables in !OpenOffice (or any other spreadsheet program). For example, to open the file {{{lh.ilf.All.table}}} in !OpenOffice, do the following : {{{ localc $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table }}} From here, you could use these stats tables to run a group analysis on the tracts. == Anisotropy and diffusivity along the trajectory of a pathway == Now take a look at the stats as a function of position along the trajectory of the same tract: {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt & }}} It will look like this: {{{ # Title Pathway Statistics # # generating_program /usr/local/freesurfer/stable6/bin/dmri_pathstats # cvs_version # cmdline /usr/local/freesurfer/stable6/bin/dmri_pathstats --intrc /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr --dtbase /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dmri/dtifit --path lh.ilf --subj elmo.2012 --out /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --outvox /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt # sysname Linux # hostname oribi # machine x86_64 # user eboyd2 # anatomy_type pathway # # subjectname elmo.2012 # pathwayname lh.ilf # # pathway start x y z AD RD MD FA AD_Avg RD_Avg MD_Avg FA_Avg 83 74 10 0.0010087 0.000830648 0.000889999 0.159232 0.00116397 0.000637616 0.000813066 0.374542 83 73 10 0.00107542 0.00076142 0.000866087 0.248381 0.00116 0.000631537 0.000807689 0.378022 83 73 11 0.00110213 0.000712833 0.000842598 0.275052 0.00116456 0.000632124 0.000809599 0.378441 83 72 11 0.00115836 0.000627003 0.000804122 0.379383 0.00118313 0.000638984 0.000820367 0.378685 83 71 11 0.00130141 0.00050243 0.000768757 0.540997 0.00120207 0.000613241 0.000809521 0.409762 83 71 12 0.00135934 0.000596314 0.000850654 0.479475 0.00122036 0.00060423 0.00080961 0.425146 83 70 12 0.00154584 0.000596379 0.000912867 0.540192 0.0012735 0.000598274 0.000823352 0.449849 83 69 12 0.0015961 0.000637489 0.000957027 0.523057 0.0013166 0.000627619 0.000857281 0.443408 83 68 13 0.00144106 0.000659587 0.000920077 0.456897 0.00131143 0.000646806 0.000868347 0.428068 84 67 13 0.00133104 0.000563732 0.000819502 0.501266 0.00126369 0.00063388 0.000843816 0.423312 84 66 14 0.00129191 0.000639349 0.000856869 0.429424 0.00120567 0.000636186 0.000826019 0.401576 84 65 14 0.00111743 0.000576156 0.000756579 0.401439 0.00120671 0.000675135 0.000852329 0.370139 84 64 15 0.000995878 0.000653256 0.000767463 0.27065 0.00115213 0.000677309 0.000835582 0.347765 84 63 15 0.00103649 0.000619989 0.000758823 0.333934 0.00120864 0.000672818 0.00085142 0.372466 84 62 16 0.00121678 0.000586189 0.000796385 0.432171 0.00128202 0.000629584 0.000847063 0.436846 84 61 16 0.00128539 0.000496937 0.000759754 0.553296 0.00133043 0.000553918 0.000812759 0.516887 84 60 16 0.00128823 0.00053104 0.000783438 0.53335 0.00134452 0.000506174 0.000785624 0.563432 84 60 17 0.00126802 0.000460207 0.000729479 0.58859 0.00133849 0.000496096 0.000776899 0.570689 84 59 17 0.00108238 0.000373869 0.000610039 0.629474 0.00133839 0.000466135 0.00075689 0.597466 84 58 17 0.00130795 0.000494729 0.000765803 0.58186 0.00140401 0.000490494 0.000795002 0.591723 84 57 18 0.00107557 0.000464563 0.000668231 0.525115 0.00141309 0.000503638 0.000806786 0.580816 84 56 18 0.00112712 0.000392396 0.000637304 0.612159 0.00142693 0.000501972 0.000810288 0.593075 84 55 19 0.00110705 0.000536791 0.000726878 0.47699 0.0014481 0.000521233 0.000830186 0.580617 84 54 19 0.00131515 0.000501804 0.000772919 0.560854 0.00149121 0.000514767 0.000840246 0.591875 84 53 19 0.00141376 0.000488109 0.000796661 0.592974 0.00151916 0.00050704 0.000844416 0.603714 84 53 20 0.00119967 0.000528667 0.000752334 0.515978 0.00151852 0.000501309 0.000840379 0.609157 84 52 20 0.00120838 0.000496335 0.000733684 0.545742 0.00151907 0.000506619 0.0008441 0.607991 84 51 21 0.0011066 0.000545991 0.000732861 0.48208 0.00145287 0.000537145 0.000842389 0.566021 83 50 21 0.00148025 0.000517207 0.000838221 0.591659 0.00143009 0.000558962 0.00084934 0.541725 83 49 21 0.00144729 0.000608569 0.000888142 0.504581 0.00143929 0.000580943 0.000867057 0.523038 83 48 21 0.00140066 0.000648933 0.000899507 0.45017 0.00146562 0.000588808 0.000881079 0.52329 82 47 22 0.00179437 0.000560765 0.000971966 0.632207 0.00152193 0.000564541 0.00088367 0.560292 82 46 22 0.00140457 0.000546434 0.00083248 0.540164 0.00148664 0.000549511 0.000861887 0.56362 81 45 22 0.00165706 0.000706209 0.00102316 0.494609 0.00148319 0.000559383 0.000867323 0.558307 80 44 23 0.00174696 0.000418429 0.000861272 0.723202 0.0014574 0.00058681 0.000877008 0.532129 80 43 23 0.00146997 0.000619092 0.000902717 0.497525 0.00140823 0.000612242 0.000877576 0.493454 79 42 23 0.00152992 0.000444628 0.000806391 0.661404 0.00139555 0.000603363 0.000867423 0.492408 78 41 23 0.00157958 0.000555758 0.000897033 0.592812 0.00138554 0.0005864 0.000852775 0.498505 78 40 24 0.00169536 0.000341967 0.000793098 0.769893 0.00136858 0.00057877 0.000842035 0.494142 77 39 24 0.00176535 0.000454217 0.000891263 0.699291 0.00136671 0.000564577 0.000831952 0.504254 76 39 24 0.00181275 0.000391344 0.000865146 0.750689 0.00138579 0.00055527 0.000832107 0.517531 76 38 25 0.00177161 0.000446334 0.000888092 0.705912 0.00138728 0.000545684 0.000826213 0.523804 75 37 25 0.00186546 0.000517308 0.000966693 0.673821 0.0013714 0.000562255 0.000831969 0.506898 74 36 25 0.00161531 0.000582424 0.000926721 0.575432 0.00135436 0.000571873 0.000832701 0.492641 74 35 25 0.00158317 0.000631253 0.000948557 0.529855 0.00134491 0.000573929 0.000830925 0.489379 73 34 25 0.00146608 0.000741013 0.000982702 0.410368 0.00134846 0.000580893 0.000836751 0.486019 72 34 25 0.00118521 0.000661327 0.000835955 0.367785 0.00134222 0.000586273 0.000838257 0.478949 72 33 25 0.00120423 0.000714338 0.000877636 0.354421 0.00133965 0.000594724 0.000843032 0.47204 # pathway end }}} This text file contains various diffusion measures, one row for each position along the trajectory of the path. The first three entries in each row are the x, y, z coordinates in native diffusion space. The next four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at that position on the maximum a posteriori path. The last four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at the same position, averaged over all sampled paths. Points are always ordered in the following way: * CST : Brainstem -> Motor cortex * UNC : Temporal -> Orbitofrontal * ILF : Temporal -> Occipital * ATR : Frontal cortex -> Thalamus * CCG : Posterior -> Anterior * CAB : Posterior -> Anterior * SLFP : Frontal -> Parietal * SLFT : Frontal -> Temporal * F-MAJOR/MINOR : Left -> Right === Converting pathstats.byvoxel.txt files to a table for group analyses === The pathstats.byvoxel.txt files will generally not contain the same number of positions (rows) for each subject because the tracts are reconstructed in each subject's native diffusion space and not in a template space. Thus they are not ready for performing group analyses yet. To combine these files from multiple subjects, interpolating the anisotropy and diffusivity values at corresponding positions along the tract for all subjects, run the following: {{{ trac-all -stat -c $TUTORIAL_DATA/diffusion_tutorial/dmrirc.tutorial }}} This will create a directory named {{{stats}}} under the main TRACULA output directory and save one table per tract per diffusion measure. In these tables, each row is a different position along the trajectory of the tract and each column is a different subject. To examine the table of average FA along the left ILF, do the following: {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.FA_Avg.txt & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.FA_Avg.txt & }}} The contents of this file will look like this: {{{ elmo.2005 elmo.2008 elmo.2012 NaN NaN 0.378069 NaN NaN 0.378492 0.41336 NaN 0.388788 0.429444 0.377014 0.4164 0.444207 0.392511 0.438363 0.452544 0.394491 0.445661 0.449954 0.388662 0.435143 0.447727 0.407221 0.426532 0.432298 0.428718 0.421008 0.419581 0.425808 0.403944 0.473702 0.40381 0.371384 0.494473 0.387531 0.353198 0.4767 0.414657 0.366791 0.506063 0.453886 0.412596 0.583493 0.519466 0.483278 0.624087 0.57846 0.549247 0.647596 0.600432 0.569196 ... }}} The first row tells you the subject name for the corresponding column of FA values. Note that for some data sets, there will be some NaN ("not a number") values in the beginning or end of the tract, because the tract has a slightly different length for each data set and the endings will not always correspond. For the purposes of a statistical analysis, you can ignore the endings, use only the data points that are not NaN, or pool together the data points from the first few or last few positions. From here, you could use these stats tables to run a group analysis on the tracts. '''Note:''' In addition to the above tables, the {{{stats}}} directory will also contain a log file for each pathway. You can examine the log file, which contains the full output of the {{{trac-all -stat}}} command, to see if any pathways were flagged as outliers. If this happens, there will be a line in the log file with {{{"Found outlier path:"}}} and the name of the subject. These are paths that were found to differ excessively from those of the other subjects. You can check this information to find out if the reconstruction of this tract failed for any subjects. For example, you can examine the log file for the left ILF by doing: {{{ gedit $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.log & }}} '''ON MACS, RUN:''' {{{ open -e $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.log & }}} === Visualizing results from statistical analyses along each pathway === The {{{trac-all -stat}}} command will also produce average paths in template space (MNI or CVS, depending on what was specified for the inter-subject registration method). These paths can be used to visualize the outputs of statistical analyses along each of the 18 tracts. Note that these average paths are only produced for visualization purposes and their existence does not imply that a voxel-based analysis was performed in template space in these coordinates. (Remember that TRACULA performs both the tractography and the extraction of anisotropy and diffusivity measures in the native diffusion space of each subject.) To view all 18 mean paths in the space of the MNI template, run the following: {{{ freeview -v $FSLDIR/data/standard/MNI152_T1_1mm_brain.nii.gz \ -w $TUTORIAL_DATA/diffusion_tutorial/stats/*.path.mean.txt }}} You should see something like this in the 3D view: {{attachment:path.waypts.jpg}} Let's say that you have performed a statistical analysis on your subjects in your statistical software of choice. If you save the p-values corresponding to each position along a tract to a simple text file, you can now display these p-values as a heat map on the corresponding mean path. (It is assumed that the number of p-values in your text file is equal to the number of points on the mean path, which is the same as the number of rows of values in the group tables produced by {{{trac-all -stat.}}} If not, display may be problematic.) To display the p-values from your hypothetical analysis on one of the mean paths, do the following: * Select the mean path of your choice on the panel in the top left of the freeview window. * From the {{{Spline color}}} menu, choose {{{Heatscale}}}. * From the {{{Scalar map}}} menu, choose {{{Load...}}} and select the text file that contains the p-values from your statistical analysis. * To hide the waypoints, which show up as small spheres, increase the {{{Spline radius}}} so that it is greater than the {{{Radius}}} of the waypoints. (In the example below, we have set the {{{Spline radius}}} to 2, while the {{{Radius}}} of the waypoints is 1.) * Set the {{{Min,}}} {{{Mid,}}} and {{{Max}}} of the heatscale to threshold the p-values as you wish. You can do this for as many of the tracts as you have performed statistical analyses on. The end result will look something like this in the 3D view: {{attachment:path.waypts.p.jpg}} |
| Line 9: | Line 333: |
| {{{ cd $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr/ less path.overall.txt less path.byvoxel.txt }}} ---- == path.overall.txt == This text file gives various diffusion measures about the specific white matter tract along its complete path. Some of the measures include: * Count of the # of Fibers in the WM tract * Tract Volume * Maximum, Minimum and Average length of fibers * Length of the center fiber * Axial Diffusivity (both average and along the center) * Radial Diffusivity (both average and along the center) * Mean Diffusivity (both average and along the center) * Fractional Anisotropy (both average and along the center) ---- {{{ # Title Pathway Statistics # # generating_program $FREESURFER_HOME/bin/dmri_pathstats # cvs_version # cmdline $FREESURFER_HOME/bin/dmri_pathstats --intrc $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr --dtbase $TUTORIAL_DATA/diffusion_tutorial/Diff001/dmri/dtifit --path lh.cst --subj Diff001 --out $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr/pathstats.overall.txt --outvox $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr/pathstats.byvoxel.txt # sysname Linux # hostname compute-0-1.local # machine x86_64 # user rspriti # anatomy_type pathway # # subjectname Diff001 # pathwayname lh.cst # Count 100 Volume 235 Len_Min 45 Len_Max 69 Len_Avg 55.3 Len_Center 53 AD_Avg 0.00108402 AD_Avg_Weight 0.00107521 AD_Avg_Center 0.00107062 RD_Avg 0.00046958 RD_Avg_Weight 0.000476065 RD_Avg_Center 0.000507009 MD_Avg 0.000674393 MD_Avg_Weight 0.000675781 MD_Avg_Center 0.000694881 FA_Avg 0.50122 FA_Avg_Weight 0.48976 FA_Avg_Center 0.456292 }}} ---- == path.byvoxel.txt == This text file gives us various diffusion measures in each voxel along the WM tract ---- {{{ # Title Pathway Statistics # # generating_program $FREESURFER_HOME/dmri_pathstats # cvs_version # cmdline $FREESURFER_HOME/dmri_pathstats --intrc $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr --dtbase $TUTORIAL_DATA/diffusion_tutorial/Diff001/dmri/dtifit --path lh.cst --subj Diff001 --out $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr/pathstats.overall.txt --outvox $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr/pathstats.byvoxel.txt # sysname Linux # hostname compute-0-1.local # machine x86_64 # user rspriti # anatomy_type pathway # # subjectname Diff001 # pathwayname lh.cst # # pathway start x y z AD RD MD FA 62 63 10 0.000637887 0.000476948 0.000530594 0.178865 62 63 11 0.0006074 0.000433505 0.00049147 0.22773 63 63 12 0.000846852 0.000610935 0.000689574 0.251636 63 64 13 0.00123916 0.00105259 0.00111478 0.107897 63 64 14 0.000936783 0.000621757 0.000726766 0.264886 64 64 15 0.000869413 0.000500918 0.00062375 0.348479 64 64 16 0.000742746 0.000432898 0.000536181 0.357518 65 64 16 0.000729103 0.000361411 0.000483975 0.42985 66 65 17 0.000645165 0.000105291 0.000285249 0.86012 67 65 18 0.00115478 0.000351219 0.000619072 0.647404 68 66 19 0.00142292 0.000690686 0.000934763 0.432432 69 66 20 0.00166259 0.000912869 0.00116278 0.363279 69 66 21 0.00145087 0.000448438 0.000782583 0.639517 70 66 22 0.00124617 0.000204701 0.000551856 0.816332 70 66 23 0.00138642 0.000415134 0.000738896 0.650657 71 66 23 0.00136252 0.000330065 0.000674217 0.721118 71 66 24 0.00134187 0.000385197 0.000704089 0.678151 71 66 25 0.00108983 0.000289931 0.000556565 0.729769 72 66 26 0.00106165 0.000357968 0.000592529 0.650427 72 66 27 0.00117242 0.000398032 0.00065616 0.619191 73 65 28 0.00130606 0.000441963 0.000729994 0.61662 73 65 29 0.00132239 0.00034928 0.000673651 0.701421 74 65 30 0.00133145 0.000391918 0.000705096 0.652609 74 65 31 0.00130265 0.000371743 0.000682045 0.665608 74 65 32 0.00126357 0.000429947 0.000707822 0.596254 75 65 33 0.00126146 0.000533254 0.000775991 0.499365 75 64 34 0.00117235 0.000465188 0.00070091 0.536046 75 64 35 0.00111144 0.000424831 0.000653701 0.560041 76 64 36 0.00128036 0.000299521 0.000626469 0.731721 76 64 37 0.00119495 0.000322181 0.000613105 0.688909 76 64 38 0.00105691 0.000334124 0.000575053 0.64994 76 63 39 0.00106784 0.000449252 0.000655449 0.514129 76 63 40 0.00113234 0.000537139 0.000735539 0.463226 76 63 41 0.000990485 0.000493054 0.000658864 0.446701 76 63 42 0.000798778 0.000477672 0.000584708 0.336071 76 62 43 0.000870652 0.000483175 0.000612334 0.362795 75 62 44 0.000878542 0.000493417 0.000621792 0.376378 75 62 45 0.000897465 0.000514559 0.000642194 0.348244 75 61 46 0.000882752 0.000537658 0.000652689 0.310019 74 61 47 0.00100985 0.00048457 0.000659663 0.436199 74 61 48 0.00104135 0.000461786 0.000654973 0.482846 74 60 49 0.000957188 0.000488566 0.000644774 0.472547 73 60 50 0.00100422 0.00055232 0.000702954 0.378698 73 60 51 0.000938077 0.000492242 0.000640854 0.429111 72 59 52 0.00104503 0.000475882 0.000665599 0.474443 72 59 53 0.00091483 0.00047261 0.000620017 0.409087 72 59 54 0.000792557 0.000632776 0.000686036 0.174565 71 59 55 0.000961412 0.000680667 0.000774249 0.243739 71 58 56 0.001061 0.000895478 0.000950651 0.133396 70 58 57 0.000862592 0.000663684 0.000729986 0.162365 70 58 58 0.00119199 0.00103056 0.00108437 0.086213 69 57 59 0.00120589 0.000980809 0.00105584 0.127098 69 57 60 0.00102806 0.000829161 0.000895461 0.141788 # pathway end }}} ---- These two files can be used to extract measures that can be further used group analysis. This can also be converted into a table using tractstats2table for doing GLM analyses using mri_glmfit or any other statistical software (SPSS, Excel, Statview etc) To convert the path.overall.txt for the above tract into a table that can be used by mri_glmfit, do the following ---- {{{ cd $TUTORIAL_DATA/diffusion_tutorial/Diff001/dpath/5cpts/priorcvs/seg14/initmni/regbbr/lh.cst_AS_avg23_cvs_bbr/ # To extract all diffusion measures into a table do the following tractstats2table |
== Study Questions == * What are the three steps involved in processing individual subjects with TRACULA and what command runs them?[[https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/DiffusionStudyQuestions#WhatarethethreestepsinvolvedinprocessingindividualsubjectswithTRACULAandwhatcommandrunsthem.3F|Answer]] * What is the configuration file? What happens if you run TRACULA without a configuration file? [[https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/DiffusionStudyQuestions#Whatistheconfigurationfile.3FWhathappensifyourunTRACULAwithoutaconfigurationfile.3F|Answer]] * Which model of diffusion does TRACULA use to reconstruct pathways from the DWI data? [[https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/DiffusionStudyQuestions#WhichmodelofdiffusiondoesTRACULAusetoreconstructpathwaysfromtheDWIdata.3F|Answer]] * What is diffusion MRI particularly sensitive to and what does this result in? How does TRACULA assess this error? [[https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial/DiffusionStudyQuestions#WhatisdiffusionMRIparticularlysensitivetoandwhatdoesthisresultin.3FHowdoesTRACULAassessthiserror.3F|Answer]] ----- [[https://surfer.nmr.mgh.harvard.edu/fswiki/Tutorials|Back to list of all tutorials]] | [[FsTutorial|Back to course page]] | [[FsTutorial/TraculaOutputs|Previous]] |
Back to list of all tutorials | Back to course page | Previous
Tract statistics
Remember...
For each new terminal that you open, you must do:
export SUBJECTS_DIR=$TUTORIAL_DATA/diffusion_recons cd $TUTORIAL_DATA/diffusion_tutorial
This section of the tutorial will teach you how to extract statistics on anisotropy and diffusivity measures for the white-matter pathways reconstructed by TRACULA. There are two types of statistics files that are created for each white-matter pathway:
- pathstats.overall.txt - This file contains measures averaged over the whole pathway
- pathstats.byvoxel.txt - This file contains measures as a function of position along the trajectory of the pathway.
For example, the statistics files for the left inferior longitudinal fasciculus (ILF) of subject elmo.2012, showing measures for the entire left ILF or along the left ILF, can be found below. Optional: use a text editor, such as gedit, or the command open -e on a Mac, to open these files.
$TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt
Anisotropy and diffusivity averaged over an entire pathway
You can view an example of the overall pathway stats by doing:
gedit $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt &
This file will look like this:
# Title Pathway Statistics # # generating_program /usr/local/freesurfer/stable6/bin/dmri_pathstats # cvs_version # cmdline /usr/local/freesurfer/stable6/bin/dmri_pathstats --intrc /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr --dtbase /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dmri/dtifit --path lh.ilf --subj elmo.2012 --out /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --outvox /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt # sysname Linux # hostname oribi # machine x86_64 # user eboyd2 # anatomy_type pathway # # subjectname elmo.2012 # pathwayname lh.ilf # Count 1500 Volume 629 Len_Min 36 Len_Max 71 Len_Avg 50.01 Len_Center 48 AD_Avg 0.00136398 AD_Avg_Weight 0.00137336 AD_Avg_Center 0.00137163 RD_Avg 0.000570444 RD_Avg_Weight 0.000568755 RD_Avg_Center 0.00056212 MD_Avg 0.000834957 MD_Avg_Weight 0.000836957 MD_Avg_Center 0.000831957 FA_Avg 0.505701 FA_Avg_Weight 0.510267 FA_Avg_Center 0.514496
This text file contains various diffusion measures, averaged over the entire white-matter tract. The measures include:
- Number of sample paths in the WM tract
- Tract volume (in voxels)
- Maximum, minimum and average length of sample paths
- Length of the highest-probability (a.k.a. maximum a posteriori) path
- Axial diffusivity (average over the entire support of the path distribution, weighted average over the entire support of the path distribution, and average over highest-probability path only)
- Radial diffusivity (as above)
- Mean diffusivity (as above)
- Fractional anisotropy (as above)
Converting pathstats.overall.txt files to a table for group analyses
Measures can be extracted from these files to be analyzed further, e.g., for tract-based group analysis. Specifically, the text files can be converted into a table using the command tractstats2table and then used for doing GLM analyses with mri_glmfit or any other statistical software (SPSS, Excel, Statview etc.)
To extract all diffusion measures for the left ILF from subject elmo.2012 into a table, do the following:
tractstats2table --inputs $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --overall --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table
The argument to the --inputs option specifies the input stats file.
The --overall option tells tractstats2table to expect the overall path stats file (stats averaged over the entire left ILF), as opposed to the stats along the trajectory of the tract (which would be specified with the --byvoxel option instead).
The argument to the --tablefile option specifies the output file.
Take a look at the resulting table file:
gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.elmo.2012.All.table &
For a group analysis, tractstats2table can be used to combine the diffusion measures in pathstats.overall.txt from multiple subjects into a single table, which can then be used for analysis with mri_glmfit or any other statistical software.
To do this, you will have to create a text file that lists the full path to every subject's pathstats.overall.txt file. An example of such a list for all 3 tutorial subjects is shown below.
$TUTORIAL_DATA/diffusion_tutorial/elmo.2005/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2008/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt
Create a text file using the command below. Copy and paste this list into it, and save the file.
gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list &
The following command will use this list to create a table with the diffusion measures from all the files listed above:
tractstats2table --load-pathstats-from-file $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list --overall --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table
The argument to the --load-pathstats-from-file option specifies the text file that contains the list of all the statistics files that will be loaded.
Take a look at the resulting table file:
gedit $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table &
Instead of extracting all measures included in the tract statistics files, we may be interested only in a few specific measures. For example, to extract only the average FA for each subject, do the following:
tractstats2table --load-pathstats-from-file $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.list --overall --only-measures FA_Avg --tablefile $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.FA_Avg.table
The argument to the --only-measures option specifies which measure we want to extract from the statistics files. Instead of FA_Avg, this could be the name of any of the measures included in pathstats.overall.txt.
OPTIONAL: You can look at these tables in OpenOffice (or any other spreadsheet program). For example, to open the file lh.ilf.All.table in OpenOffice, do the following :
localc $TUTORIAL_DATA/diffusion_tutorial/lh.ilf.All.table
From here, you could use these stats tables to run a group analysis on the tracts.
Anisotropy and diffusivity along the trajectory of a pathway
Now take a look at the stats as a function of position along the trajectory of the same tract:
gedit $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt &
It will look like this:
# Title Pathway Statistics # # generating_program /usr/local/freesurfer/stable6/bin/dmri_pathstats # cvs_version # cmdline /usr/local/freesurfer/stable6/bin/dmri_pathstats --intrc /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr --dtbase /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dmri/dtifit --path lh.ilf --subj elmo.2012 --out /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.overall.txt --outvox /autofs/space/rama_001/users/eboyd2/FStutorial_edits/diffusion_tutorial/elmo.2012/dpath/lh.ilf_AS_avg33_mni_bbr/pathstats.byvoxel.txt # sysname Linux # hostname oribi # machine x86_64 # user eboyd2 # anatomy_type pathway # # subjectname elmo.2012 # pathwayname lh.ilf # # pathway start x y z AD RD MD FA AD_Avg RD_Avg MD_Avg FA_Avg 83 74 10 0.0010087 0.000830648 0.000889999 0.159232 0.00116397 0.000637616 0.000813066 0.374542 83 73 10 0.00107542 0.00076142 0.000866087 0.248381 0.00116 0.000631537 0.000807689 0.378022 83 73 11 0.00110213 0.000712833 0.000842598 0.275052 0.00116456 0.000632124 0.000809599 0.378441 83 72 11 0.00115836 0.000627003 0.000804122 0.379383 0.00118313 0.000638984 0.000820367 0.378685 83 71 11 0.00130141 0.00050243 0.000768757 0.540997 0.00120207 0.000613241 0.000809521 0.409762 83 71 12 0.00135934 0.000596314 0.000850654 0.479475 0.00122036 0.00060423 0.00080961 0.425146 83 70 12 0.00154584 0.000596379 0.000912867 0.540192 0.0012735 0.000598274 0.000823352 0.449849 83 69 12 0.0015961 0.000637489 0.000957027 0.523057 0.0013166 0.000627619 0.000857281 0.443408 83 68 13 0.00144106 0.000659587 0.000920077 0.456897 0.00131143 0.000646806 0.000868347 0.428068 84 67 13 0.00133104 0.000563732 0.000819502 0.501266 0.00126369 0.00063388 0.000843816 0.423312 84 66 14 0.00129191 0.000639349 0.000856869 0.429424 0.00120567 0.000636186 0.000826019 0.401576 84 65 14 0.00111743 0.000576156 0.000756579 0.401439 0.00120671 0.000675135 0.000852329 0.370139 84 64 15 0.000995878 0.000653256 0.000767463 0.27065 0.00115213 0.000677309 0.000835582 0.347765 84 63 15 0.00103649 0.000619989 0.000758823 0.333934 0.00120864 0.000672818 0.00085142 0.372466 84 62 16 0.00121678 0.000586189 0.000796385 0.432171 0.00128202 0.000629584 0.000847063 0.436846 84 61 16 0.00128539 0.000496937 0.000759754 0.553296 0.00133043 0.000553918 0.000812759 0.516887 84 60 16 0.00128823 0.00053104 0.000783438 0.53335 0.00134452 0.000506174 0.000785624 0.563432 84 60 17 0.00126802 0.000460207 0.000729479 0.58859 0.00133849 0.000496096 0.000776899 0.570689 84 59 17 0.00108238 0.000373869 0.000610039 0.629474 0.00133839 0.000466135 0.00075689 0.597466 84 58 17 0.00130795 0.000494729 0.000765803 0.58186 0.00140401 0.000490494 0.000795002 0.591723 84 57 18 0.00107557 0.000464563 0.000668231 0.525115 0.00141309 0.000503638 0.000806786 0.580816 84 56 18 0.00112712 0.000392396 0.000637304 0.612159 0.00142693 0.000501972 0.000810288 0.593075 84 55 19 0.00110705 0.000536791 0.000726878 0.47699 0.0014481 0.000521233 0.000830186 0.580617 84 54 19 0.00131515 0.000501804 0.000772919 0.560854 0.00149121 0.000514767 0.000840246 0.591875 84 53 19 0.00141376 0.000488109 0.000796661 0.592974 0.00151916 0.00050704 0.000844416 0.603714 84 53 20 0.00119967 0.000528667 0.000752334 0.515978 0.00151852 0.000501309 0.000840379 0.609157 84 52 20 0.00120838 0.000496335 0.000733684 0.545742 0.00151907 0.000506619 0.0008441 0.607991 84 51 21 0.0011066 0.000545991 0.000732861 0.48208 0.00145287 0.000537145 0.000842389 0.566021 83 50 21 0.00148025 0.000517207 0.000838221 0.591659 0.00143009 0.000558962 0.00084934 0.541725 83 49 21 0.00144729 0.000608569 0.000888142 0.504581 0.00143929 0.000580943 0.000867057 0.523038 83 48 21 0.00140066 0.000648933 0.000899507 0.45017 0.00146562 0.000588808 0.000881079 0.52329 82 47 22 0.00179437 0.000560765 0.000971966 0.632207 0.00152193 0.000564541 0.00088367 0.560292 82 46 22 0.00140457 0.000546434 0.00083248 0.540164 0.00148664 0.000549511 0.000861887 0.56362 81 45 22 0.00165706 0.000706209 0.00102316 0.494609 0.00148319 0.000559383 0.000867323 0.558307 80 44 23 0.00174696 0.000418429 0.000861272 0.723202 0.0014574 0.00058681 0.000877008 0.532129 80 43 23 0.00146997 0.000619092 0.000902717 0.497525 0.00140823 0.000612242 0.000877576 0.493454 79 42 23 0.00152992 0.000444628 0.000806391 0.661404 0.00139555 0.000603363 0.000867423 0.492408 78 41 23 0.00157958 0.000555758 0.000897033 0.592812 0.00138554 0.0005864 0.000852775 0.498505 78 40 24 0.00169536 0.000341967 0.000793098 0.769893 0.00136858 0.00057877 0.000842035 0.494142 77 39 24 0.00176535 0.000454217 0.000891263 0.699291 0.00136671 0.000564577 0.000831952 0.504254 76 39 24 0.00181275 0.000391344 0.000865146 0.750689 0.00138579 0.00055527 0.000832107 0.517531 76 38 25 0.00177161 0.000446334 0.000888092 0.705912 0.00138728 0.000545684 0.000826213 0.523804 75 37 25 0.00186546 0.000517308 0.000966693 0.673821 0.0013714 0.000562255 0.000831969 0.506898 74 36 25 0.00161531 0.000582424 0.000926721 0.575432 0.00135436 0.000571873 0.000832701 0.492641 74 35 25 0.00158317 0.000631253 0.000948557 0.529855 0.00134491 0.000573929 0.000830925 0.489379 73 34 25 0.00146608 0.000741013 0.000982702 0.410368 0.00134846 0.000580893 0.000836751 0.486019 72 34 25 0.00118521 0.000661327 0.000835955 0.367785 0.00134222 0.000586273 0.000838257 0.478949 72 33 25 0.00120423 0.000714338 0.000877636 0.354421 0.00133965 0.000594724 0.000843032 0.47204 # pathway end
This text file contains various diffusion measures, one row for each position along the trajectory of the path. The first three entries in each row are the x, y, z coordinates in native diffusion space. The next four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at that position on the maximum a posteriori path. The last four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at the same position, averaged over all sampled paths.
Points are always ordered in the following way:
CST : Brainstem -> Motor cortex
UNC : Temporal -> Orbitofrontal
ILF : Temporal -> Occipital
ATR : Frontal cortex -> Thalamus
CCG : Posterior -> Anterior
CAB : Posterior -> Anterior
SLFP : Frontal -> Parietal
SLFT : Frontal -> Temporal
F-MAJOR/MINOR : Left -> Right
Converting pathstats.byvoxel.txt files to a table for group analyses
The pathstats.byvoxel.txt files will generally not contain the same number of positions (rows) for each subject because the tracts are reconstructed in each subject's native diffusion space and not in a template space. Thus they are not ready for performing group analyses yet. To combine these files from multiple subjects, interpolating the anisotropy and diffusivity values at corresponding positions along the tract for all subjects, run the following:
trac-all -stat -c $TUTORIAL_DATA/diffusion_tutorial/dmrirc.tutorial
This will create a directory named stats under the main TRACULA output directory and save one table per tract per diffusion measure. In these tables, each row is a different position along the trajectory of the tract and each column is a different subject.
To examine the table of average FA along the left ILF, do the following:
gedit $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.FA_Avg.txt &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.FA_Avg.txt &
The contents of this file will look like this:
elmo.2005 elmo.2008 elmo.2012 NaN NaN 0.378069 NaN NaN 0.378492 0.41336 NaN 0.388788 0.429444 0.377014 0.4164 0.444207 0.392511 0.438363 0.452544 0.394491 0.445661 0.449954 0.388662 0.435143 0.447727 0.407221 0.426532 0.432298 0.428718 0.421008 0.419581 0.425808 0.403944 0.473702 0.40381 0.371384 0.494473 0.387531 0.353198 0.4767 0.414657 0.366791 0.506063 0.453886 0.412596 0.583493 0.519466 0.483278 0.624087 0.57846 0.549247 0.647596 0.600432 0.569196 ...
The first row tells you the subject name for the corresponding column of FA values. Note that for some data sets, there will be some NaN ("not a number") values in the beginning or end of the tract, because the tract has a slightly different length for each data set and the endings will not always correspond. For the purposes of a statistical analysis, you can ignore the endings, use only the data points that are not NaN, or pool together the data points from the first few or last few positions.
From here, you could use these stats tables to run a group analysis on the tracts.
Note: In addition to the above tables, the stats directory will also contain a log file for each pathway. You can examine the log file, which contains the full output of the trac-all -stat command, to see if any pathways were flagged as outliers. If this happens, there will be a line in the log file with "Found outlier path:" and the name of the subject. These are paths that were found to differ excessively from those of the other subjects. You can check this information to find out if the reconstruction of this tract failed for any subjects.
For example, you can examine the log file for the left ILF by doing:
gedit $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.log &
ON MACS, RUN:
open -e $TUTORIAL_DATA/diffusion_tutorial/stats/lh.ilf_AS.avg33_mni_bbr.log &
Visualizing results from statistical analyses along each pathway
The trac-all -stat command will also produce average paths in template space (MNI or CVS, depending on what was specified for the inter-subject registration method). These paths can be used to visualize the outputs of statistical analyses along each of the 18 tracts. Note that these average paths are only produced for visualization purposes and their existence does not imply that a voxel-based analysis was performed in template space in these coordinates. (Remember that TRACULA performs both the tractography and the extraction of anisotropy and diffusivity measures in the native diffusion space of each subject.)
To view all 18 mean paths in the space of the MNI template, run the following:
freeview -v $FSLDIR/data/standard/MNI152_T1_1mm_brain.nii.gz \
-w $TUTORIAL_DATA/diffusion_tutorial/stats/*.path.mean.txtYou should see something like this in the 3D view:
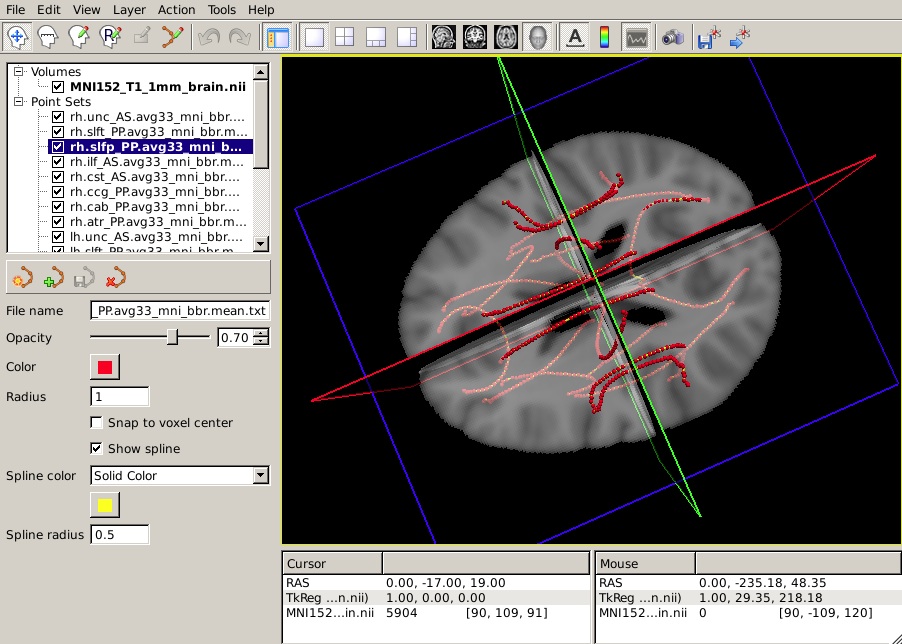
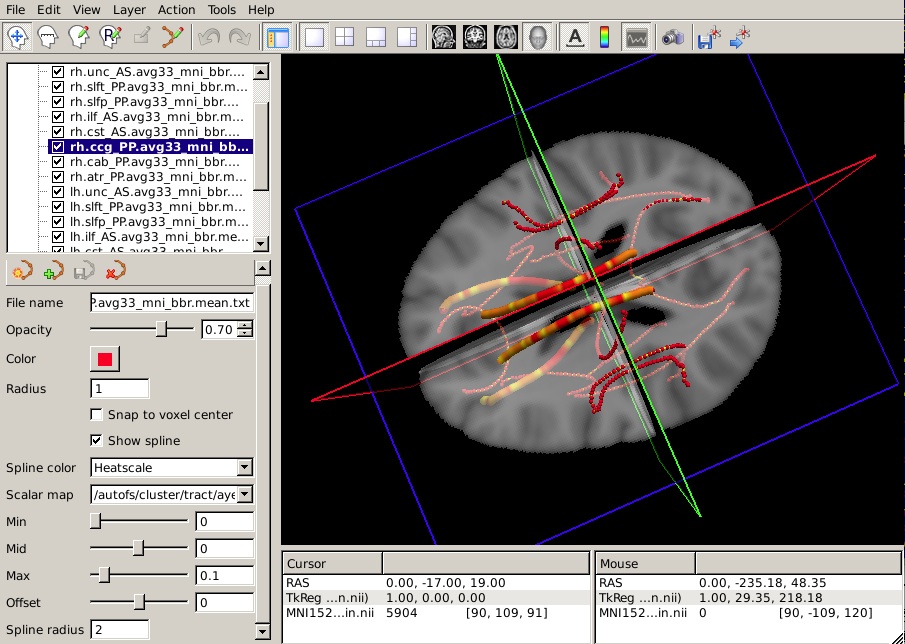
Let's say that you have performed a statistical analysis on your subjects in your statistical software of choice. If you save the p-values corresponding to each position along a tract to a simple text file, you can now display these p-values as a heat map on the corresponding mean path. (It is assumed that the number of p-values in your text file is equal to the number of points on the mean path, which is the same as the number of rows of values in the group tables produced by trac-all -stat. If not, display may be problematic.)
To display the p-values from your hypothetical analysis on one of the mean paths, do the following:
- Select the mean path of your choice on the panel in the top left of the freeview window.
From the Spline color menu, choose Heatscale.
From the Scalar map menu, choose Load... and select the text file that contains the p-values from your statistical analysis.
To hide the waypoints, which show up as small spheres, increase the Spline radius so that it is greater than the Radius of the waypoints. (In the example below, we have set the Spline radius to 2, while the Radius of the waypoints is 1.)
Set the Min, Mid, and Max of the heatscale to threshold the p-values as you wish.
You can do this for as many of the tracts as you have performed statistical analyses on. The end result will look something like this in the 3D view:
Study Questions
What are the three steps involved in processing individual subjects with TRACULA and what command runs them?Answer
What is the configuration file? What happens if you run TRACULA without a configuration file? Answer
Which model of diffusion does TRACULA use to reconstruct pathways from the DWI data? Answer
What is diffusion MRI particularly sensitive to and what does this result in? How does TRACULA assess this error? Answer
Back to list of all tutorials | Back to course page | Previous
