Back to list of all tutorials | Back to course page | Previous
Tract statistics
Please bear with us while we update the tutorial data set for the new version of TRACULA. The new tutorial goes beyond what was possible with the tutorial data set that was previously available for download. For now, please ignore any references to the tutorial data and try to follow along using your own data. Thank you for your patience. |
Remember...
For each new terminal that you open, you must do:
export SUBJECTS_DIR=$TUTORIAL_DATA/diffusion_tutorial/fs cd $TUTORIAL_DATA/diffusion_tutorial
This section of the tutorial will teach you how to extract statistics on anisotropy and diffusivity measures for the white-matter pathways reconstructed by TRACULA. There are two types of statistics files that are created for each white-matter pathway:
pathstats.overall.txt - This file contains diffusion measures averaged over the whole pathway.
pathstats.byvoxel.txt - This file contains diffusion measures averaged at consecutive cross-sections of the pathway, resulting in an along-tract profile of each diffusion measure. This type of analysis is referred to as pointwise assessment of streamline tractography attributes (PASTA).
These are plain text files that you can open with a text editor, such as gedit on Linux, or with the command open -e on a Mac. They are saved in the TRACULA output directory of each individual tract.
Anisotropy and diffusivity averaged over an entire pathway
Examine the whole-tract statistics for the left inferior longitudinal fasciculus (ILF) of subject1, by runnning:
cat trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.overall.txt
This file will look like this:
# Title Pathway Statistics # # generating_program /usr/local/freesurfer/7.2.0-beta/bin/dmri_pathstats # cvs_version ayendiki-local # cmdline /usr/local/freesurfer/7.2.0-beta/bin/dmri_pathstats --intrc /space/erebus/1/users/data/trc/subject1/dpath/lh.ilf_avg16_syn_bbr --dtbase /space/erebus/1/users/data/trc/subject1/dmri/dtifit --path lh.ilf --subj subject1 --invox path.ref.txt --out /space/erebus/1/users/data/trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.overall.txt --outvox /space/erebus/1/users/data/trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.byvoxel.txt # sysname Linux # hostname compute-0-124.nmr.mgh.harvard.edu # machine x86_64 # user ayendiki # anatomy_type pathway # # subjectname subject # pathwayname lh.ilf # Count 1500 Volume 1996 Len_Min 53 Len_Max 129 Len_Avg 79.9787 Len_Center 89 AD_Avg 0.00120044 AD_Avg_Weight 0.00121209 AD_Avg_Center 0.00123318 RD_Avg 0.000467543 RD_Avg_Weight 0.000458132 RD_Avg_Center 0.000549716 MD_Avg 0.000711842 MD_Avg_Weight 0.000709453 MD_Avg_Center 0.000777538 FA_Avg 0.540167 FA_Avg_Weight 0.553683 FA_Avg_Center 0.477498
This text file contains various diffusion measures, averaged over the entire pathway (in this example, the left ILF). The measures include:
- Number of sample paths drawn from the probability distribution of the pathway (Count)
- Volume of the probability distribution of the pathway (in voxels)
- Maximum, minimum, and average length of the sample paths (in voxels)
- Length of the highest-probability (a.k.a. maximum a posteriori) path
- Axial diffusivity (average over the entire support of the path distribution, weighted average over the entire support of the path distribution, and average over highest-probability path only)
- Radial diffusivity (as above)
- Mean diffusivity (as above)
- Fractional anisotropy (as above)
Combining pathstats.overall.txt files for group analyses
Measures can be extracted from these files and concatenated across subjects, e.g., for group analysis. Specifically, the text files can be converted into a table using the command tractstats2table and then used for doing GLM analyses with mri_glmfit or any other statistical software (SPSS, Excel, Statview etc.)
Before using tractstats2table, you have to create a text file that lists the full path to the pathstats.overall.txt files from all subjects that you want to include in your analysis. For example, if you wanted to analyze whole-tract measures from the left ILF of 3 subjects, you would create a text file that looks like the following:
$TUTORIAL_DATA/diffusion_tutorial/trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/trc/subject2/dpath/lh.ilf_avg16_syn_bbr/pathstats.overall.txt $TUTORIAL_DATA/diffusion_tutorial/trc/subject3/dpath/lh.ilf_avg16_syn_bbr/pathstats.overall.txt
Create a text file called lh.ilf.list using the command below. Copy and paste the list above into this file, then save the file and exit the editor.
gedit lh.ilf.list &
ON MACS, RUN:
open -e lh.ilf.list &
The following command will use this list to create a table with the diffusion measures from all the subjects listed above:
tractstats2table --load-pathstats-from-file lh.ilf.list --overall --tablefile lh.ilf.All.table
The argument to the --load-pathstats-from-file option specifies the text file that contains the list of all the statistics files that will be combined.
Take a look at the resulting table file:
gedit lh.ilf.All.table &
ON MACS, RUN:
open -e h.ilf.All.table &
Instead of extracting all measures included in the pathstats.overall.txt files, we are usually interested only in a few specific measures. For example, to extract only the average FA for each subject, do the following:
tractstats2table --load-pathstats-from-file $lh.ilf.list --overall --only-measures FA_Avg --tablefile lh.ilf.FA_Avg.table
The argument to the --only-measures option specifies which measure we want to extract from the statistics files. Instead of FA_Avg, this could be the name of any of the measures included in pathstats.overall.txt.
OPTIONAL: You can look at these tables in OpenOffice (or any other spreadsheet program). For example, to open the file lh.ilf.All.table in OpenOffice, do the following :
localc lh.ilf.All.table
You can then use these stats tables as input to mri_glmfit, to run perform group analysis.
Anisotropy and diffusivity along the trajectory of a pathway
Now examine the along-tract (PASTA) statistics for the left ILF, by running:
cat trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.byvoxel.txt
# Title Pathway Statistics # # generating_program /usr/local/freesurfer/7.2.0-beta/bin/dmri_pathstats # cvs_version ayendiki-local # cmdline /usr/local/freesurfer/7.2.0-beta/bin/dmri_pathstats --intrc /space/erebus/1/users/data/trc/subject1/dpath/lh.ilf_avg16_syn_bbr --dtbase /space/erebus/1/users/data/trc/subject1/dmri/dtifit --path lh.ilf --subj subject1 --invox path.ref.txt --out /space/erebus/1/users/data/trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.overall.txt --outvox /space/erebus/1/users/data/trc/subject1/dpath/lh.ilf_avg16_syn_bbr/pathstats.byvoxel.txt # sysname Linux # hostname compute-0-124.nmr.mgh.harvard.edu # machine x86_64 # user ayendiki # anatomy_type pathway # # subjectname subject1 # pathwayname lh.ilf # # pathway start x y z AD RD MD FA AD_Avg RD_Avg MD_Avg FA_Avg 97 78 16 0.000867977 0.000725794 0.000773188 0.108892 0.00107168 0.000599572 0.000756942 0.363722 97 78 16 0.000867977 0.000725794 0.000773188 0.108892 0.00107168 0.000599572 0.000756942 0.363722 97 77 16 0.000832794 0.000712196 0.000752395 0.098711 0.00106892 0.000599915 0.000756251 0.363217 97 77 16 0.000832794 0.000712196 0.000752395 0.098711 0.00106892 0.000599915 0.000756251 0.363217 96 77 17 0.000778157 0.000713259 0.000734892 0.0524794 0.00106822 0.000599456 0.000755712 0.364947 ... 85 30 46 0.00161654 0.000434083 0.000828234 0.688725 0.00121171 0.000469266 0.000716745 0.540934 85 29 45 0.00159702 0.000373003 0.00078101 0.732022 0.00120427 0.000464151 0.000710859 0.54041 84 29 45 0.00149992 0.000433867 0.000789218 0.665445 0.0012176 0.000470298 0.000719398 0.53854 84 29 45 0.00149992 0.000433867 0.000789218 0.665445 0.0012176 0.000470298 0.000719398 0.53854 84 29 45 0.00149992 0.000433867 0.000789218 0.665445 0.0012176 0.000470298 0.000719398 0.53854 # pathway end
This text file contains various diffusion measures, one row for each position along the trajectory of the path. The first three entries in each row are the x, y, z coordinates in native diffusion space. The next four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at that position on the maximum a posteriori path. The last four entries are the axial diffusivity, radial diffusivity, mean diffusivity, and fractional anisotropy at the same position, averaged over all sampled paths.
Points are ordered according to the following convention (see also along-tract plots):
- For pathways with a right-left orientation:
- If the pathway is inter-hemispheric: From right to left
- If the pathway is intra-hemispheric: From medial to lateral
- For pathways with an anterior-posterior orientation: From anterior to posterior
- For pathways with a superior-inferior orientation: From superior to inferior
Combining pathstats.byvoxel.txt files for group analyses
You can combine the pathstats.byvoxel.txt files from multiple subjects, to use as input for along-tract group analysis, by running the following:
trac-all -stat -c $TUTORIAL_DATA/diffusion_tutorial/dmrirc.tutorial
This will create a directory named stats under the main TRACULA output directory and save one table per tract per diffusion measure. In these tables, each row is a different position along the trajectory of the tract and each column is a different subject.
To examine the table of average FA along the left ILF, do the following:
gedit trc/stats/lh.ilf.avg16_syn_bbr.FA_Avg.txt &
ON MACS, RUN:
open -e trc/stats/lh.ilf.avg16_syn_bbr.FA_Avg.txt &
The contents of this file will look like this:
subject1 subject2 subject3 0.429444 0.377014 0.4164 0.444207 0.392511 0.438363 0.452544 0.394491 0.445661 0.449954 0.388662 0.435143 0.447727 0.407221 0.426532 0.432298 0.428718 0.421008 0.419581 0.425808 0.403944 0.473702 0.40381 0.371384 0.494473 0.387531 0.353198 0.4767 0.414657 0.366791 0.506063 0.453886 0.412596 0.583493 0.519466 0.483278 0.624087 0.57846 0.549247 0.647596 0.600432 0.569196 ...
The first row tells you the subject names for the corresponding columns of FA values.
You can use these stats tables to run a group analysis on the tracts.
Note: In addition to the above tables, the stats directory will also contain a log file for each pathway. You can examine the log file, which contains the full output of the trac-all -stat command, to see if any pathways were flagged as outliers. If this happens, there will be a line in the log file with "Found outlier path:" and the name of the subject. These are paths that were found to differ excessively from those of the other subjects. You can check this information to find out if the reconstruction of this tract failed for any subjects.
For example, you can examine the log file for the left ILF by doing:
gedit trc/stats/lh.ilf.avg16_syn_bbr.log &
ON MACS, RUN:
open -e trc/stats/lh.ilf.avg16_syn_bbr.log &
Visualizing results from statistical analyses along each pathway
By default, TRACULA uses the mean of its manually annotated training streamlines to determine the positions along each of the 42 pathways where diffusion measures will be projected. These mean streamlines are saved in template space (the template that is the target of the inter-subject registration) and mapped to each individual subject after the individual subjects pathways are reconstructed, to extract the along-tract measures. Note that these mean paths are only used to determine how each pathway will sliced up into into cross-sections where along-tract measures will be averaged, and to ensure an equal number of cross-sections for all subjects.
To view all 42 mean paths in template space, run the following:
freeview -v $FREESURFER_HOME/trctrain/hcp/MGH35_HCP_FA_template.nii.gz \
-t $FREESURFER_HOME/trctrain/hcp/syn/*.mean.trkYou should see something like this in the 3D view:
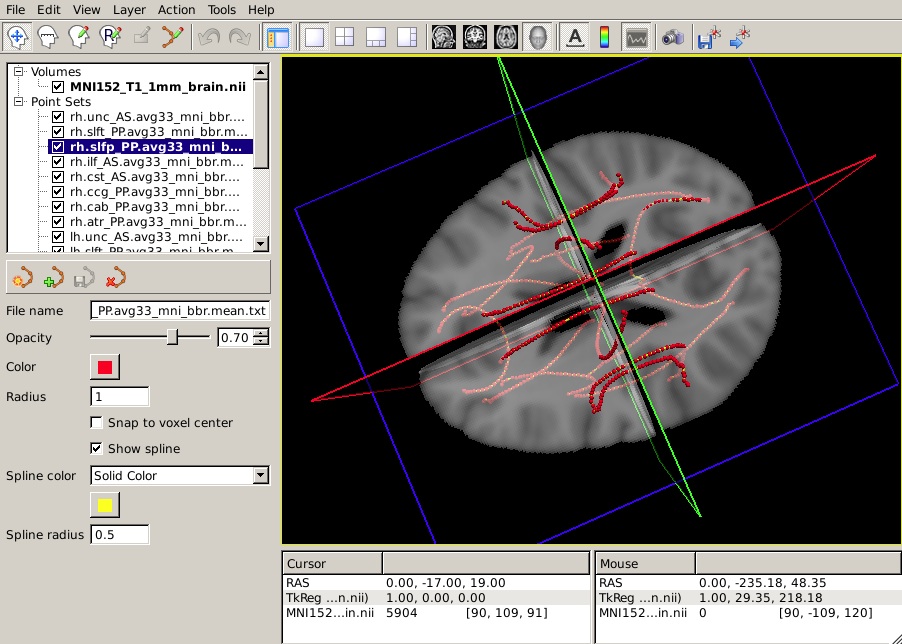
The following information is based on hypothetical statistical data. The text file of p-values is not available so do not expect your end result to look identical to this image.
Let's say that you have performed a statistical analysis on your subjects in your statistical software of choice. If you save the p-values corresponding to each position along a tract to a simple text file, you can now display these p-values as a heat map on the corresponding mean path. (It is assumed that the number of p-values in your text file is equal to the number of points on the mean path, which is the same as the number of rows of values in the group tables produced by trac-all -stat. If not, display may be problematic.)
To display the p-values from your hypothetical analysis on one of the mean paths, do the following:
Select the mean path of your choice on the panel in the top left of the freeview window. On the left menu, click on Show spline.
From the Spline color menu, choose Heatscale.
From the Scalar map menu, choose Load... and select the text file that contains the p-values from your statistical analysis.
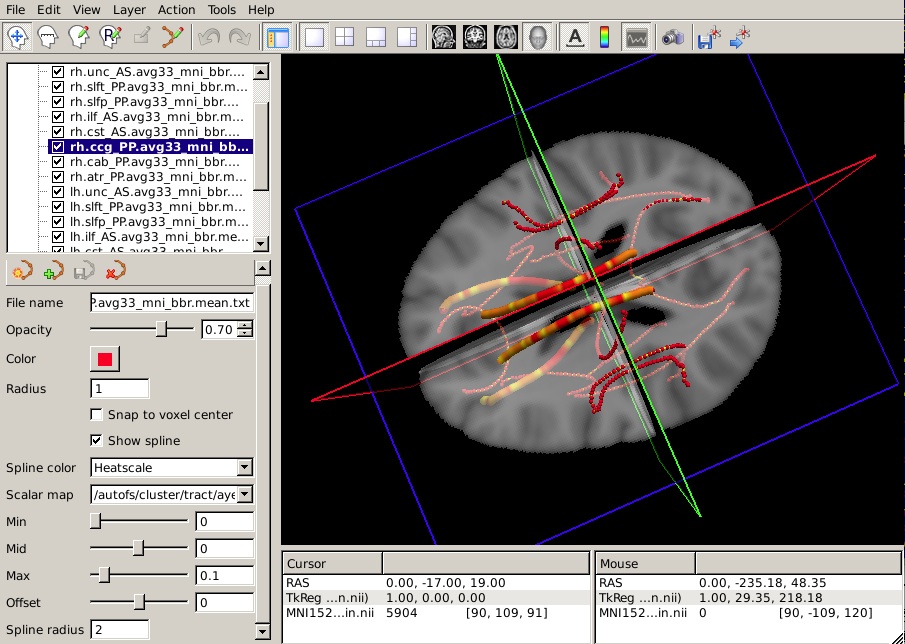
To hide the waypoints, which show up as small spheres, increase the Spline radius so that it is greater than the Radius of the waypoints. (In the example below, we have set the Spline radius to 2, while the Radius of the waypoints is 1.)
Set the Min, Mid, and Max of the heatscale to threshold the p-values as you wish.
You can do this for as many of the tracts as you have performed statistical analyses on. The end result will look something like this in the 3D view:
Summary
By the end of this page, you should know how to:
- Extract statistics (anisotropy and diffusivity measures), either averaged over an entire white-matter pathway, or as a function of position along the trajectory of the pathway
- Combine statistics into tables for group analysis
- Perform whole-tract and along-tract statistical analysis
- Visualize the outputs of along-tract statistical analysis in freeview
Quiz
You can test your knowledge of this tutorial by clicking here for a quiz!
Back to list of all tutorials | Back to course page | Previous
