|
Size: 9950
Comment:
|
Size: 8200
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 14: | Line 14: |
| A preprint of the manuscript describing the atlas and segmentation method used in this module can be found [[https://www.biorxiv.org/content/10.1101/2024.02.05.579016v1|on bioRxiv.]] | Relevant publications: |
| Line 16: | Line 16: |
| "A probabilistic histological atlas of the human brain for MRI segmentation", Casamitjana et al., accepted. [[https://www.biorxiv.org/content/10.1101/2024.02.05.579016v1|Preprint available here.]] <<BR>> "Fast segmentation with the NextBrain histological atlas", Puonti et al., submitted. [[https://www.biorxiv.org/content/10.1101/2025.09.22.673638v1|Preprint available here.]] <<BR>> <<BR>> |
|
| Line 20: | Line 28: |
| 4. Usage ('full' Bayesian version, '''not recommended''') 5. Frequently asked questions (FAQ) |
4. SynthMorph version 5. Legacy mode 6. Frequently asked questions (FAQ) |
| Line 25: | Line 34: |
| This module uses our new probabilistic atlas of the human brain to segment 333 distinct ROIs per hemisphere on in vivo scans. Segmentation relies on a Bayesian algorithm and is thus robust against changes in MRIi pulse sequence (e.g., T1-weighted, T2-weighted, FLAIR, etc). Sample slices of the atlas and the segmentation of the sample subject "bert" are shown below: | This module uses NextBrain, our new probabilistic atlas of the human brain, to segment ~300 distinct ROIs per hemisphere on in vivo or ex vivo scans (including single hemispheres). Segmentation relies on a Bayesian algorithm and is thus robust against changes in MRI pulse sequence (e.g., T1-weighted, T2-weighted, FLAIR, etc). Sample slices of the atlas and the segmentation of the sample subject "bert" are shown below: |
| Line 29: | Line 38: |
| The first time you run this module, it will prompt you to download the atlas. Follow the instructions on the screen to obtain the files. | The first time you run this module, it will prompt you to download the atlas and machine learning model files. Follow the instructions on the screen to obtain the files. |
| Line 32: | Line 41: |
| === 3. Usage (faster versions) === | === 3. Usage === |
| Line 34: | Line 43: |
| '''We just released these, so please bear with us (and definitely let us know!) if the code is buggy or crashes.''' Our original implementation (still available for compatibility and reproducibility, see Section 4 below) used a "proper" Bayesian inference algorithm that makes segmentation incredibly slow (tens of hours) unless a GPU with pretty large RAM is available. Since December 2024, we distribute two faster versions, where the atlas deformation is pre-computed and then kept constant during the optimization, such that we only need to run the EM algorithm once for the Gaussian parameters. Segmentation of 1mm isotropic scans takes about 20 minutes on a CPU with 8 cores (and just a few minutes on a GPU). Segmentation of ex vivo scans with 0.25mm isotropic resolution takes about 2 hours on the same CPU. There are two such fast versions. One uses [[https://arxiv.org/abs/2404.01249|FireANTs (Jena et al)]] to register to the target scan a synthetic cartoon derived from the atlas. This version actively tries to fit smaller structures and subregions. The command line is: |
The default version of the algorithm is compatible with in vivo and ex vivo brains and relies on [[https://arxiv.org/abs/2404.01249|FireANTs (Jena et al)]] for the registration part of the algorithm. The command line is: |
| Line 51: | Line 46: |
| mri_histo_atlas_segment_fireants INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS [BF_MODE] | mri_histo_atlas_segment_fireants INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS [MODE] |
| Line 62: | Line 57: |
| ''BF_MODE'' (optional): bias field mode: dct (default), polynomial, or hybrid (try polynomial/hybrid if default doesn't do well) | ''MODE'' (optional): type of scan: invivo (default), exvivo, cerebrum (ex vivo without brainstem or cerebellum), hemi (ex vivo with single cerebral hemisphere). |
| Line 64: | Line 59: |
| For example, you can segment bert with a GPU and 8 CPU threads with: | The output directory will contain the following files: |
| Line 66: | Line 61: |
| {{{ mri_histo_atlas_segment $SUBJECTS_DIR/bert/mri/orig.mgz /path/to/output/bert_histo_atlas_segmentation/ 1 8 }}} |
''seg.[left/right].nii.gz'': segmentation of left/right hemisphere |
| Line 70: | Line 63: |
| In the output directory, you will find: | ''lut.txt'': the lookup table to visualize seg.[left/right].nii.gz, for convenience |
| Line 72: | Line 65: |
| ''seg.[left/right].mgz'': NextBrain segmentation of the left and right hemisphere, respectively. | ''vols.[left/right].csv'': files with volumes of the brain regions segmented by the atlas, in CSV format. |
| Line 74: | Line 67: |
| ''vols.[left/right].csv'': CSV spreadsheet with the volumes of the different ROIs, computed from the posteriors (soft segmentations). | ''supersynth.nii.gz'': segmentation of the scan at the whole structure level (from the foundation model) |
| Line 76: | Line 69: |
| ''seg.[left/right].rgb.mgz'': color version of the soft segmentation (use "Display as RBG map" option in Freeview). ''synthseg.mgz'': [[https://surfer.nmr.mgh.harvard.edu/fswiki/SynthSeg|SynthSeg]] segmentation of the input (used in preprocessing, to make cartoon, and to initialize GMM parameters). ''SynthSeg_volumes.csv'': coarse volumes [[https://surfer.nmr.mgh.harvard.edu/fswiki/SynthSeg|SynthSeg]] segmentation of the input (used in preprocessing, to make cartoon, and to initialize GMM parameters). ''atlas_reg.nii.gz'': [[https://surfer.nmr.mgh.harvard.edu/fswiki/EasyReg|EasyReg]] registration to MNI space, used in preprocessing. ''lut.txt'': FreeSurfer lookup table mapping label indices to brain anatomy. You need it when visualizing the NextBrain segmentations with Freeview. ''atlas_nonlinear_reg.[left/right].nii.gz'': registered cartoon images. Useful for debugging, if the segmentation fails. The other fast version uses [[https://arxiv.org/abs/2404.01249|SynthMorph (Hoffmann et al., Imaging Neuroscience, 2024)]], a neural network for image registration. This version is even faster (because of SynthMorph and because it runs both hemispheres in one go) and does OK for 1mm isotropic scans. However, SynthMorph relies heavily on fitting the boundaries of whole structures and does not the map smaller regions as well (e.g., thalamic nuclei). Therefore, we do not recommend it for images with resolution better than 1mm isotropic (e.g., ex vivo scans). {{{ mri_histo_atlas_segment_fast INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS [BF_MODE] }}} The outputs are very similar to that of mri_histo_atlas_segment_fireants. |
''supersynth.vols.csv'': volumes estimated by foundation model |
| Line 107: | Line 73: |
| === 4. Usage ('full' Bayesian version; not recommended as it is very slow) === | === 4. SynthMorph version === There is a version of the code that estimates the registration with [[https://direct.mit.edu/imag/article/doi/10.1162/imag_a_00197/121549/Anatomy-aware-and-acquisition-agnostic-joint|SynthMorph (Hoffmann et al., Imaging Neuroscience, 2024)]], rather than FireANTs. This version is very good for 1mm isotropic in vivo scans. However, SynthMorph relies heavily on fitting the boundaries of whole structures and does not the map smaller regions (e.g., thalamic nuclei) as well as the FireANTs version. Therefore, we do not recommend it for images with resolution better than 1mm isotropic. Also, it does not support ex vivo scans, cerebra, or single hemispheres. the command is: {{{ mri_histo_atlas_segment_synthmorph INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS }}} The input arguments and structure of the output directory are very similar to those of mri_histo_atlas_segment_fireants. <<BR>> === 5. Legacy mode === |
| Line 111: | Line 88: |
| Our original implementation (still available for compatibility and reproducibilityw) used a "proper" Bayesian inference algorithm that makes segmentation incredibly slow (tens of hours) unless a GPU with pretty large RAM is available. |
|
| Line 113: | Line 95: |
| mri_histo_atlas_segment INPUT_SCAN OUTPUT_DIRECTORY ATLAS_MODE GPU THREADS [BF_MODE] [GMM_MODE] | mri_histo_atlas_segment_fullbayesian INPUT_SCAN OUTPUT_DIRECTORY ATLAS_MODE GPU THREADS [BF_MODE] [GMM_MODE] |
| Line 147: | Line 129: |
| <<BR>> | |
| Line 149: | Line 132: |
| === 6. Frequently asked questions (FAQ) === | |
| Line 150: | Line 134: |
| * '''Do I need a GPU?''' | |
| Line 151: | Line 136: |
=== 5. Frequently asked questions (FAQ) === * '''The output of mri_histo_atlas_segment_fireants looks generally OK but is off in some areas; can I do something about it?''' This is particularly important for high-resolution ex vivo brains. There is still hope! If you open the SynthSeg segmentation in the results directory and it is off in the offending area, you can manually edit it (which doesn't take long since it is always at 1mm resolution) and re-run mri_histo_atlas_segment_fireants with the same output directory. |
Certainly no! The code should run in less than half an hour on any semi-modern workstation, if you allocate enough threads (or about two hours for an ex vivo scan at 0.25mm resolution). |
| Line 162: | Line 141: |
* '''Do I need a GPU for the faster versions?''' Certainly no! The code should run in less than half an hour on any semi-modern workstation, if you allocate enough threads (or about two hours for an ex vivo scan at 0.25mm resolution). |
|
| Line 180: | Line 154: |
| Note that, in in dev versions newer than August 23 2024, these files are located under data_full and data_simplified (one directory per atlas / protocol). |
Bayesian Segmentation with Histological Atlas "NextBrain"
Visit the homepage of the NextBrain project for further information on this atlas.
Author: Juan Eugenio Iglesias
E-mail: jiglesiasgonzalez [at] mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
Relevant publications:
"A probabilistic histological atlas of the human brain for MRI segmentation", Casamitjana et al., accepted. Preprint available here.
"Fast segmentation with the NextBrain histological atlas", Puonti et al., submitted. Preprint available here.
Contents
- General Description
- Installation
- Usage
SynthMorph version
- Legacy mode
- Frequently asked questions (FAQ)
1. General Description
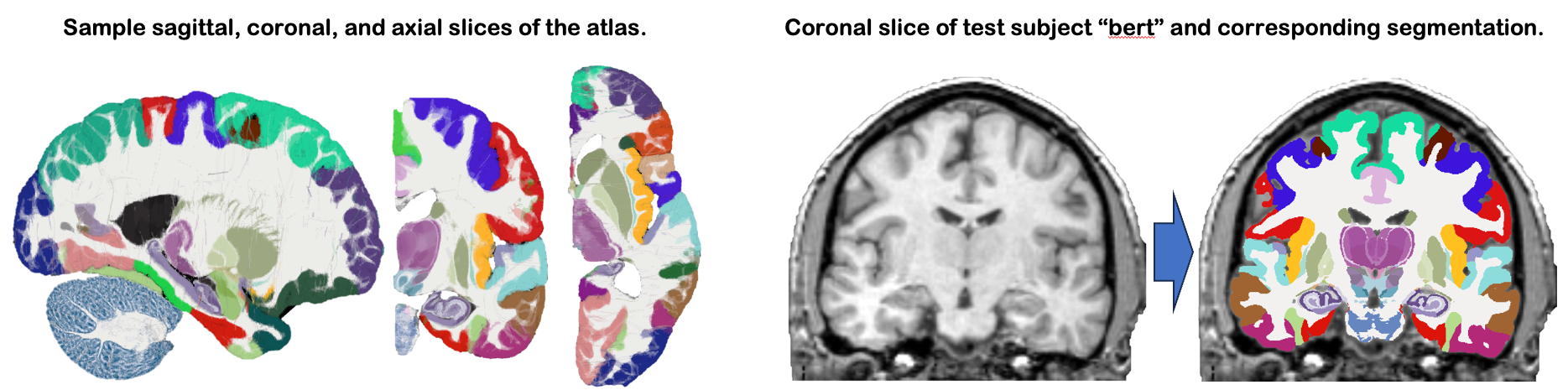
This module uses NextBrain, our new probabilistic atlas of the human brain, to segment ~300 distinct ROIs per hemisphere on in vivo or ex vivo scans (including single hemispheres). Segmentation relies on a Bayesian algorithm and is thus robust against changes in MRI pulse sequence (e.g., T1-weighted, T2-weighted, FLAIR, etc). Sample slices of the atlas and the segmentation of the sample subject "bert" are shown below:
2. Installation
The first time you run this module, it will prompt you to download the atlas and machine learning model files. Follow the instructions on the screen to obtain the files.
3. Usage
The default version of the algorithm is compatible with in vivo and ex vivo brains and relies on FireANTs (Jena et al) for the registration part of the algorithm. The command line is:
mri_histo_atlas_segment_fireants INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS [MODE]
INPUT_SCAN: scan to process, in mgz or nii(.gz) format.
OUTPUT_DIRECTORY: directory where segmentations, volume files, etc, will be created (more on this below).
GPU: set to 1 to use the GPU (faster, but you many run out of memory!).
THREADS: number of CPU threads used by the code (set to -1 to use all available threads).
MODE (optional): type of scan: invivo (default), exvivo, cerebrum (ex vivo without brainstem or cerebellum), hemi (ex vivo with single cerebral hemisphere).
The output directory will contain the following files:
seg.[left/right].nii.gz: segmentation of left/right hemisphere
lut.txt: the lookup table to visualize seg.[left/right].nii.gz, for convenience
vols.[left/right].csv: files with volumes of the brain regions segmented by the atlas, in CSV format.
supersynth.nii.gz: segmentation of the scan at the whole structure level (from the foundation model)
supersynth.vols.csv: volumes estimated by foundation model
4. SynthMorph version
There is a version of the code that estimates the registration with SynthMorph (Hoffmann et al., Imaging Neuroscience, 2024), rather than FireANTs. This version is very good for 1mm isotropic in vivo scans. However, SynthMorph relies heavily on fitting the boundaries of whole structures and does not the map smaller regions (e.g., thalamic nuclei) as well as the FireANTs version. Therefore, we do not recommend it for images with resolution better than 1mm isotropic. Also, it does not support ex vivo scans, cerebra, or single hemispheres. the command is:
mri_histo_atlas_segment_synthmorph INPUT_SCAN OUTPUT_DIRECTORY GPU THREADS
The input arguments and structure of the output directory are very similar to those of mri_histo_atlas_segment_fireants.
5. Legacy mode
We discourage users from using this version, which we only keep for compatibility and reproducibility.
Our original implementation (still available for compatibility and reproducibilityw) used a "proper" Bayesian inference algorithm that makes segmentation incredibly slow (tens of hours) unless a GPU with pretty large RAM is available.
To segment a brain MRI scan,
mri_histo_atlas_segment_fullbayesian INPUT_SCAN OUTPUT_DIRECTORY ATLAS_MODE GPU THREADS [BF_MODE] [GMM_MODE]
where:
INPUT_SCAN: scan to process, in mgz or nii(.gz) format.
OUTPUT_DIRECTORY: directory where segmentations, volume files, etc, will be created (more on this below).
ATLAS_MODE : full (atlas with all 333 labels) or simplified (simpler brainstem protocol used by fast versions; recommended)
GPU: set to 1 to use the GPU (we highly recommend using a GPU to run this module; without a GPU, running this module on a single scan can take tens of hours). The GPU requirements depend on the image but are about 24GB of memory.
THREADS: number of CPU threads used by the code (set to -1 to use all available threads).
BF_MODE (optional): bias field mode: dct (default), polynomial, or hybrid.
GMM_MODE (optional): gaussian mixture model (GMM) mode: must be 1mm unless you define your own (see FAQ below).
In the output directory, you will find:
bf_corrected.mgz: a bias field corrected version of the input scan.
SynthSeg.mgz: SynthSeg segmentation of the input (which we use in preprocessing and to initialize Gaussian parameters).
MNI_registration.mgz: EasyReg registration to MNI space, use in preprocessing.
seg_[left/right].mgz: segmentation into 333 ROIs of the left and right hemisphere, respectively.
vols_[left/right].csv: CSV spreadsheet with the volumes of the different ROIs, computed from the posteriors (soft segmentations).
lookup_table.txt: FreeSurfer lookup table mapping label indices to brain anatomy. You need it when visualizing the segmentations with Freeview.
6. Frequently asked questions (FAQ)
Do I need a GPU?
Certainly no! The code should run in less than half an hour on any semi-modern workstation, if you allocate enough threads (or about two hours for an ex vivo scan at 0.25mm resolution).
Do I really need a GPU for the 'full' Bayesian version?
Technically, no. In practice, yes. On a modern GPU, the code runs in an hour or less. On the CPU, it depends on the number of threads, but it can easily take a whole day or more.
What do the BF_MODE and GMM_MODE arguments do in the full Bayesian version?
You should not need to touch these, but the BF_MODE changes the set of basis functions for bias field correction and you could potentially try tinkering with it if the bias field correction fails (i.e., if bf_corrected.mgz has noticeable bias). GMM_MODE allows you to change the grouping of ROIs into tissue classes (advanced mode!). The GMM model is crucial as it determines how different brain regions are grouped into tissue types for the purpose of image intensity modeling. This is specified though a set of files that should be found under 'data' in the atlas directory:
data/gmm_components_[GMM_MODE].yaml: defines tissue classes and specificies the number of components of the corresponding GMM
data/combined_aseg_labels_[GMM_MODE].yaml defines the labels that belong to each tissue class
data/combined_atlas_labels_[GMM_MODE].yaml defines FreeSurfer ('aseg') labels that are used to initialize the parameters of each class.
We distribute a GMM_MODE named "1mm" that we have used in our experiments, and which is the default mode of the code. If you want to use your own model, you will need to create another triplet of files of your own (use the 1mm version as template).
