|
Size: 1782
Comment:
|
Size: 1788
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 31: | Line 31: |
| <<BR>> {{attachment:3D.png||height="300"}} <<BR>><<BR>> | <<BR>> {{attachment:3D_small.png||height="500"}} <<BR>><<BR>> |
This page is readable only by those in the LcnGroup and CmetGroup.
Samseg (cross-sectional, longitudinal, MS lesions)
This functionality is available in FreeSurfer 7
Author: Koen Van Leemput
E-mail: koen [at] nmr.mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use these tools in your analysis, please cite:
Cross-sectional: Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. O Puonti, JE Iglesias, K Van Leemput. Neuroimage, 143, 235-249, 2016.
- Longitudinal:
- MS lesions:
See also: ThalamicNuclei, HippocampalSubfieldsAndNucleiOfAmygdala, BrainstemSubstructures
1. General Description
This tool
The following figure illustrates
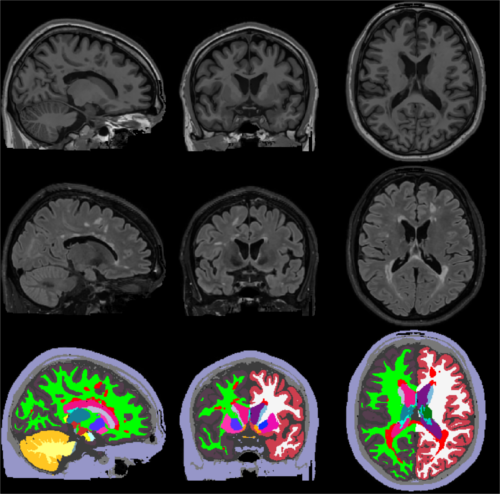
2. Installation
The first time you run this module, it will prompt you to install Tensorflow. Simply follow the instructions in the screen to install the CPU or GPU version.
If you have a compatible GPU, you can install the GPU version for faster processing, but this requires installing libraries (GPU driver, Cuda, CuDNN). These libraries are generally required for a GPU, and are not specific for this tool. In fact you may have already installed them. In this case you can directly use this tool without taking any further actions, as the code will automatically run on your GPU.
