|
Size: 2198
Comment:
|
Size: 2347
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| = Samseg = | = Samseg (cross-sectional, longitudinal, MS lesions) = |
| Line 6: | Line 6: |
| == Introduction == | '''''This functionality is available in [[https://surfer.nmr.mgh.harvard.edu/fswiki/ReleaseNotes|FreeSurfer 7]]''''' but with gradual improvements in the development version. |
| Line 8: | Line 8: |
| '''SAMseg''' is the general name of the processing stream intended to replace the subcortical segmentation stream in FreeSurfer, including some of the intensity correction and skull stripping steps preceding mri_ca_register/label. | ''Author: Koen Van Leemput'' |
| Line 10: | Line 10: |
| SAMseg is an acronym for Sequence-Adaptive Model for Segmentation, and is based on the work of Oula Puonti, Juan Eugenio Iglesias and Koen Van Leemput: [[http://orbit.dtu.dk/files/127427974/Fast_and_sequence_adaptive.pdf|Fast and Sequence-Adaptive Whole-Brain Segmentation Using Parametric Bayesian Modeling]] | ''E-mail: koen [at] nmr.mgh.harvard.edu'' |
| Line 12: | Line 12: |
| Work on Samseg is driven by three distinct grant aims: | ''Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu'' |
| Line 14: | Line 14: |
| 1. Doug Greve, FreeSurfer Maintenance R01: Replacement of the current FreeSurfer segmentation stream with one that is faster, and most importantly accepts multimodal data. This grant is also funding acquisition of subject data to compose a new multi-modal, labeled atlas, in addition to algorithm development. | If you use these tools in your analysis, please cite: |
| Line 16: | Line 16: |
| 2. CorticoMetrics, Morphometry Phase II SBIR: Replacement of the current FS seg stream with one that is faster, but still based solely on T1-weighted input, and using the existing FS atlas. As such, it is a technical milestone of the Maintenance grant, but one conducted by CorticoMetrics in its grant with the sole focus on speed of execution relative to current freesurfer. | * Cross-sectional: [[http://nmr.mgh.harvard.edu/~koen/PuontiNI2016.pdf|Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling]]. O. Puonti, J.E. Iglesias, K. Van Leemput. Neuroimage, 143, 235-249, 2016. |
| Line 18: | Line 18: |
| 3. Andre van der Kouwe, FS on-the-scanner (future submission): The ability to generate structure segmentations in near-real-time on an MRI scanner (or GPU'd extension), possibly accepting lower-res data and less-than current FS quality segmentations (in the name of identifying FOV of structures to acquire). | * Longitudinal: [[https://arxiv.org/pdf/2008.05117.pdf|A Longitudinal Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis]]. S. Cerri, A. Hoopes, D.N. Greve, M. Mühlau, K. Van Leemput. International Workshop on Machine Learning in Neuroimaging, 2020. |
| Line 20: | Line 20: |
| This page is intended to document the multiple fronts of work composing these aims. | * MS lesions: [[https://arxiv.org/pdf/2005.05135|A Contrast-Adaptive Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis]]. S. Cerri, O. Puonti, D.S. Meier, J. Wuerfel, M. Mühlau, H.R. Siebner, K. Van Leemput. 2020. |
| Line 22: | Line 22: |
| == Project Fronts == | See also: ThalamicNuclei, HippocampalSubfieldsAndNucleiOfAmygdala, BrainstemSubstructures |
| Line 24: | Line 24: |
| * Evaluate existing Samseg tools - See [[SamsegEvaluationMarch2017]] * Bring Samseg tools into FS repository * Replacement of SPM registration tool - See [[SamsegAffine]] * Testing of Samseg-T1 against FS aseg - See [[SamsegAsegTesting]] * Subject acquisition |
<<BR>> === 1. General Description === This tool The following figure illustrates <<BR>> {{attachment:3D_small.png||height="500"}} <<BR>><<BR>> |
| Line 31: | Line 34: |
| === 2. Installation === The first time you run this module, it will prompt you to install Tensorflow. Simply follow the instructions in the screen to install the CPU or GPU version. If you have a compatible GPU, you can install the GPU version for faster processing, but this requires installing libraries (GPU driver, Cuda, CuDNN). These libraries are generally required for a GPU, and are not specific for this tool. In fact you may have already installed them. In this case you can directly use this tool without taking any further actions, as the code will automatically run on your GPU. |
|
| Line 33: | Line 41: |
| <<BR>> === 3. Usage === |
This page is readable only by those in the LcnGroup and CmetGroup.
Samseg (cross-sectional, longitudinal, MS lesions)
This functionality is available in FreeSurfer 7 but with gradual improvements in the development version.
Author: Koen Van Leemput
E-mail: koen [at] nmr.mgh.harvard.edu
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use these tools in your analysis, please cite:
Cross-sectional: Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. O. Puonti, J.E. Iglesias, K. Van Leemput. Neuroimage, 143, 235-249, 2016.
Longitudinal: A Longitudinal Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis. S. Cerri, A. Hoopes, D.N. Greve, M. Mühlau, K. Van Leemput. International Workshop on Machine Learning in Neuroimaging, 2020.
MS lesions: A Contrast-Adaptive Method for Simultaneous Whole-Brain and Lesion Segmentation in Multiple Sclerosis. S. Cerri, O. Puonti, D.S. Meier, J. Wuerfel, M. Mühlau, H.R. Siebner, K. Van Leemput. 2020.
See also: ThalamicNuclei, HippocampalSubfieldsAndNucleiOfAmygdala, BrainstemSubstructures
1. General Description
This tool
The following figure illustrates
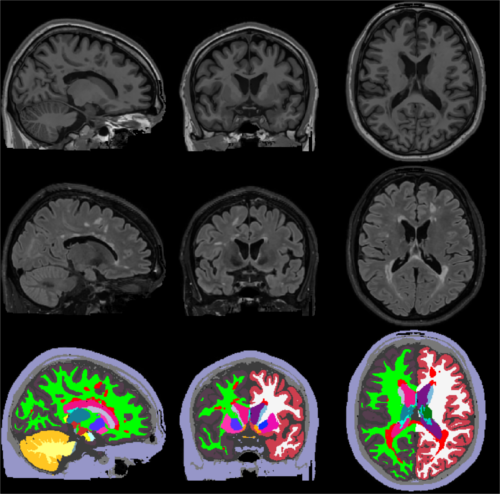
2. Installation
The first time you run this module, it will prompt you to install Tensorflow. Simply follow the instructions in the screen to install the CPU or GPU version.
If you have a compatible GPU, you can install the GPU version for faster processing, but this requires installing libraries (GPU driver, Cuda, CuDNN). These libraries are generally required for a GPU, and are not specific for this tool. In fact you may have already installed them. In this case you can directly use this tool without taking any further actions, as the code will automatically run on your GPU.
