|
Size: 1237
Comment: new Slicer starter-page
|
← Revision 28 as of 2009-04-22 16:25:11 ⇥
Size: 2440
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| [[http://www.slicer.org|3D Slicer]] is a software program for visualization, registration, segmentation, and quantification of medical data. It is developed as a collaboration between the MIT AI Lab and the Surgical Planning Lab at Brigham & Women's Hospital. The 3D Slicer homepage is [[http://www.slicer.org|http://www.slicer.org]]. | |
| Line 3: | Line 4: |
| 3D Slicer is a software program for visualization, registration, segmentation, and quantification of medical data. It is devloped as a collaboration between the MIT AI Lab and the Surgical Planning Lab at Brigham & Women's Hospital. The 3D Slicer homepage is [http://www.slicer.org http://www.slicer.org]. | == MGH NMR Center Installation == Within the MGH NMR Center, 3D Slicer is already installed on the network for both the Linux and Mac OS (PowerPC and Intel) platforms. You start 3D Slicer by typing: |
| Line 5: | Line 7: |
| == NMR Center availability == | {{{ slicer3 }}} Notes: |
| Line 7: | Line 12: |
| Within the MGH NMR Center, 3D Slicer is already installed on the network for both the Linux and Mac OS (PowerPC and Intel) platforms. You start 3D Slicer v2.6 by typing: | * A command named 'slicer' exists in the FSL toolset, and is completely unrelated to the 3D Slicer application, so remember not to confuse the two. * 'slicer3' is a wrapper-script found in /usr/pubsw/bin, which starts 3D Slicer for the platform you are using. Older and alpha versions of 3D Slicer are found in /usr/pubsw/packages/slicer. The one considered the most stable is found in /usr/pubsw/packages/slicer/current. * The old version 2 of 3D Slicer is also available. This command is used to start it: |
| Line 12: | Line 19: |
| == Download == Remember that you do not need to install 3D Slicer if you are connected to the NMR Center network (see above). |
|
| Line 13: | Line 22: |
| Notes: * A command named 'slicer' already exists in the FSL tools, and is completely unrelated to the 3D Slicer application, so remember not to confuse the two. * Soon, version 3 of 3D Slicer will become available. When this is available, this command will be used to start it: |
If you want to install 3D Slicer on your non-networked computer, go to the [[http://www.slicer.org/slicerWiki/index.php/Slicer:Slicer2.6_Getting_Started|Slicer2.6_Getting_Started]] page and follow the simple instructions for your machine. |
| Line 17: | Line 24: |
| {{{ slicer3 }}} == Download == If you want to install 3D Slicer on your non-networked computer, follow the 'Get the Software' link on the [http://www.slicer.org 3D Slicer home page]. Or, if you are feeling brave, you can go [http://www.na-mic.org/Slicer/Download/Release/ directly to the binary download site] and get the tarball. |
== Usage == To learn more about using Slicer, refer to the [[http://www.na-mic.org/Wiki/index.php/Slicer:Workshops:User_Training_101|Slicer_User_Training_101]] page which contains self-guided tutorials and sample data for Slicer2.6. Specifically, the [[http://www.na-mic.org/Wiki/images/4/4e/SlicerTraining6_vtkFreeSurferReaders.ppt|Slicer Training 6: vtkFreeSurferReaders Module tutorial|&do=get]] will teach you how to visualize Freesurfer generated subcortical segmentations and cortical parcellations both as color coded shading and as 3D models (see screenshots below). |
| Line 27: | Line 30: |
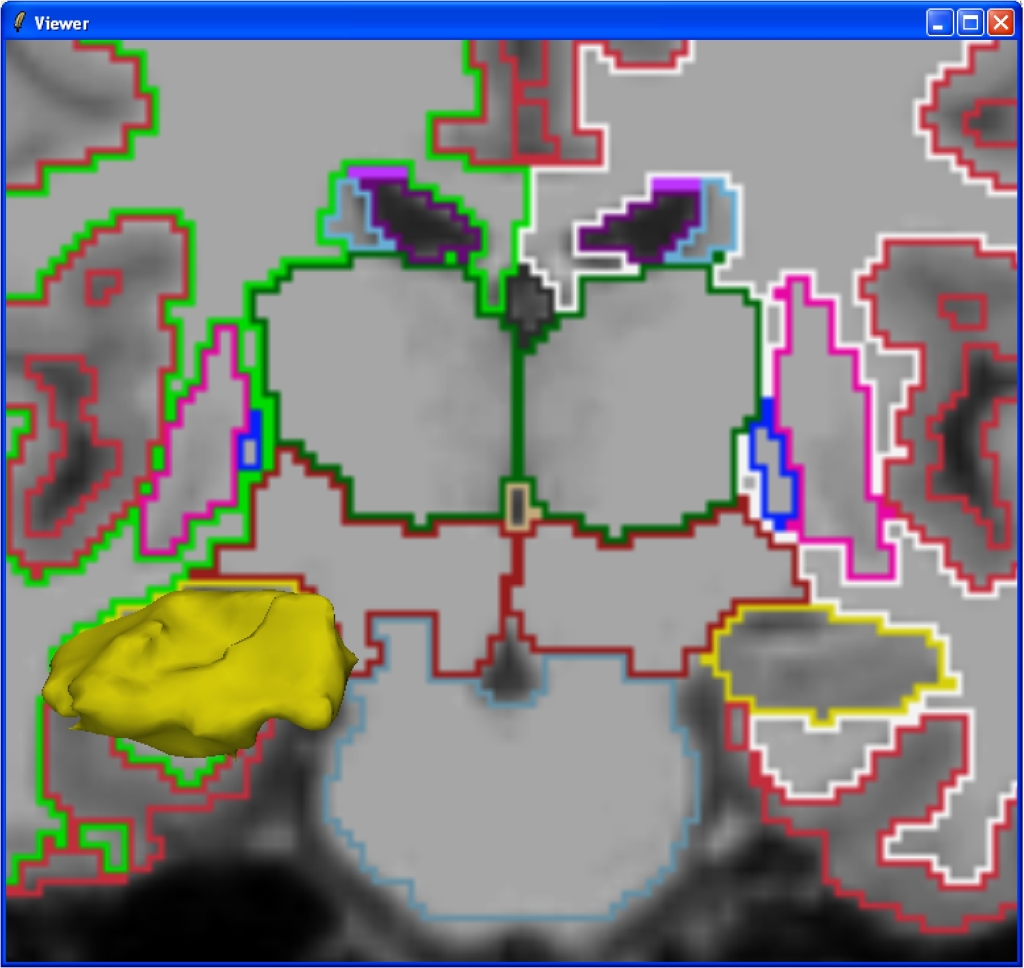
| == Screenshots == Below are screenshots of 3D Slicer (slicer v2.6) making use of the vtkFreeSurferReaders module to display subcortical segmentation (aseg.mgz) slices and selected structure volumes: |
|
| Line 28: | Line 33: |
| {{attachment:FreesurferSlicer1.jpg}} Detail of the hippocampus: {{attachment:FreesurferSlicer2.jpg}} |
3D Slicer
3D Slicer is a software program for visualization, registration, segmentation, and quantification of medical data. It is developed as a collaboration between the MIT AI Lab and the Surgical Planning Lab at Brigham & Women's Hospital. The 3D Slicer homepage is http://www.slicer.org.
MGH NMR Center Installation
Within the MGH NMR Center, 3D Slicer is already installed on the network for both the Linux and Mac OS (PowerPC and Intel) platforms. You start 3D Slicer by typing:
slicer3
Notes:
- A command named 'slicer' exists in the FSL toolset, and is completely unrelated to the 3D Slicer application, so remember not to confuse the two.
- 'slicer3' is a wrapper-script found in /usr/pubsw/bin, which starts 3D Slicer for the platform you are using. Older and alpha versions of 3D Slicer are found in /usr/pubsw/packages/slicer. The one considered the most stable is found in /usr/pubsw/packages/slicer/current.
- The old version 2 of 3D Slicer is also available. This command is used to start it:
slicer2
Download
Remember that you do not need to install 3D Slicer if you are connected to the NMR Center network (see above).
If you want to install 3D Slicer on your non-networked computer, go to the Slicer2.6_Getting_Started page and follow the simple instructions for your machine.
Usage
To learn more about using Slicer, refer to the Slicer_User_Training_101 page which contains self-guided tutorials and sample data for Slicer2.6. Specifically, the Slicer Training 6: vtkFreeSurferReaders Module tutorial will teach you how to visualize Freesurfer generated subcortical segmentations and cortical parcellations both as color coded shading and as 3D models (see screenshots below).
Work Flows
See the SlicerWorkFlows page.
Screenshots
Below are screenshots of 3D Slicer (slicer v2.6) making use of the vtkFreeSurferReaders module to display subcortical segmentation (aseg.mgz) slices and selected structure volumes:
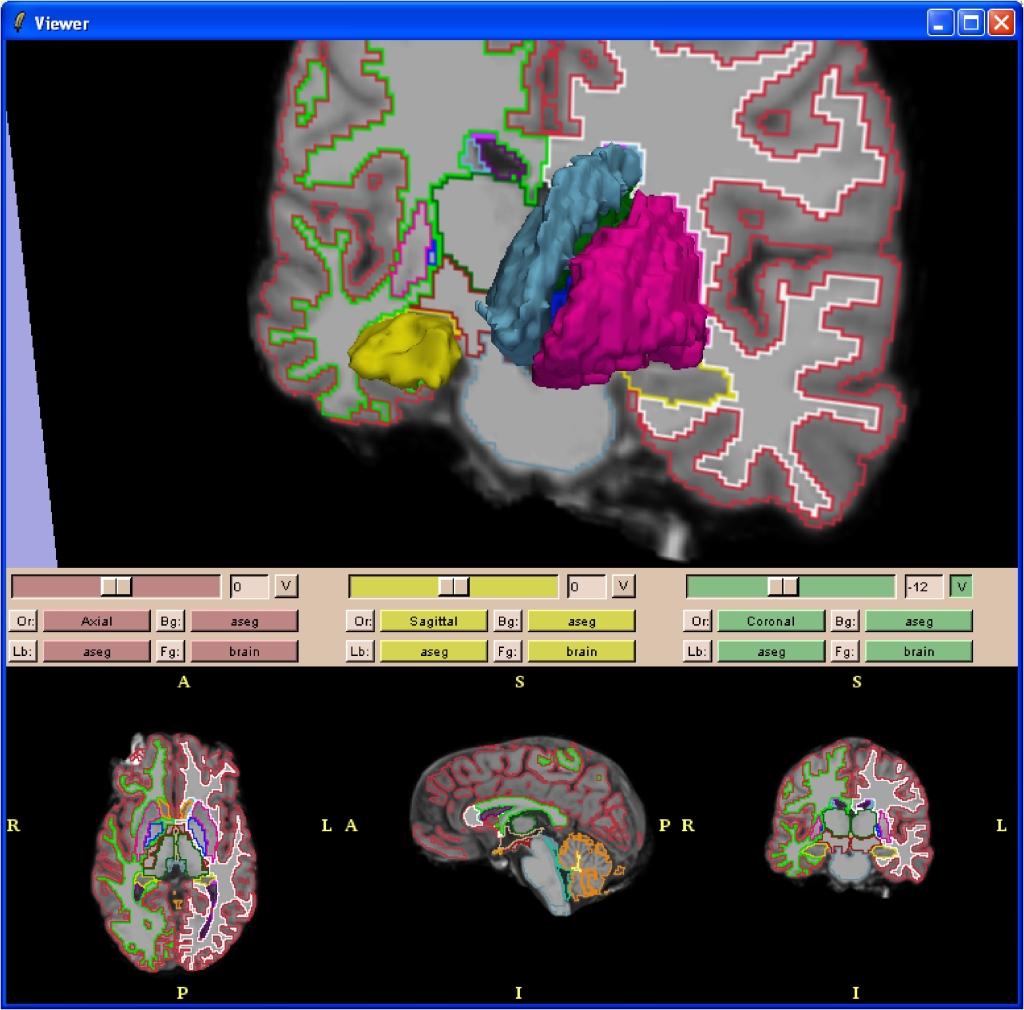
Detail of the hippocampus: