Contents
mri_TumorSynth
mri_TumorSynth - To segment the healthy brain tissue and tumor in MR scans with tumor with support for multi-sequence MR inputs.
Author/s: Jiaming Wu & Ferran Prados Carrasco
E-mail: jiaming.wu.20 [at] ucl.ac.uk or f.carrasco [at] ucl.ac.uk
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
General Description
mri_TumorSynth is a convolutional neural network-based tool designed for integrated segmentation of healthy brain tissue and brain tumors in MR scans containing tumors. It operates out-of-the-box with minimal user input, supporting clinical and research workflows focused on brain tumor analysis.
Key features of mri_TumorSynth:
- Dual-mode segmentation: Whole-tumor + healthy tissue (--wholetumor) and fine-grained inner tumor substructures (--innertumor) following BraTS criteria.
- Robust to variable scan qualities: Works with clinical MR sequences (T1, T1CE, T2, FLAIR, etc.) of different resolutions and contrasts.
- Multi-sequence input support: accepts combined T1CE+T2+FLAIR+... (or other MRI modalities) inputs for improved segmentation accuracy (see Usage section).
FreeSurfer-compatible: Output labels align with FreeSurfer’s anatomical classification for seamless integration with downstream pipelines (e.g., volume analysis, visualization).
- Efficient processing: Runs on both GPU (10s per scan) and CPU (3 minutes per scan) with minimal computational overhead.
- Enhanced robustness to low-resolution scans (≤2mm isotropic): Internal resampling and augmentation reduce error in clinical datasets.
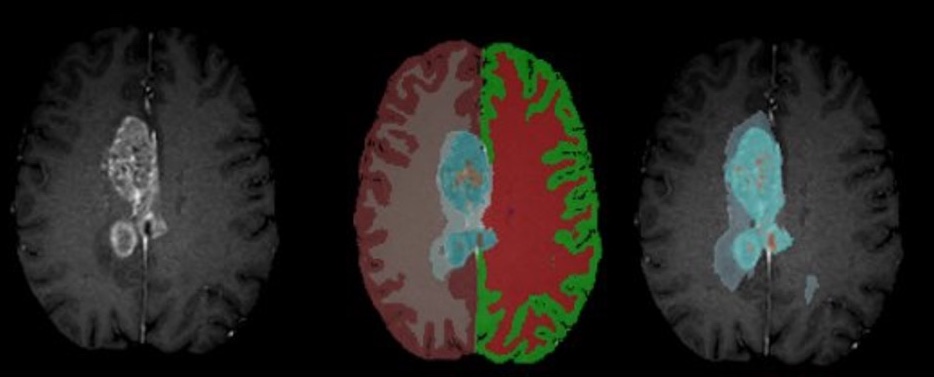
Figure: Example segmentation results – Input T1CE scan (left), healthy tissue + whole tumor mask (middle), inner tumor substructures (right).
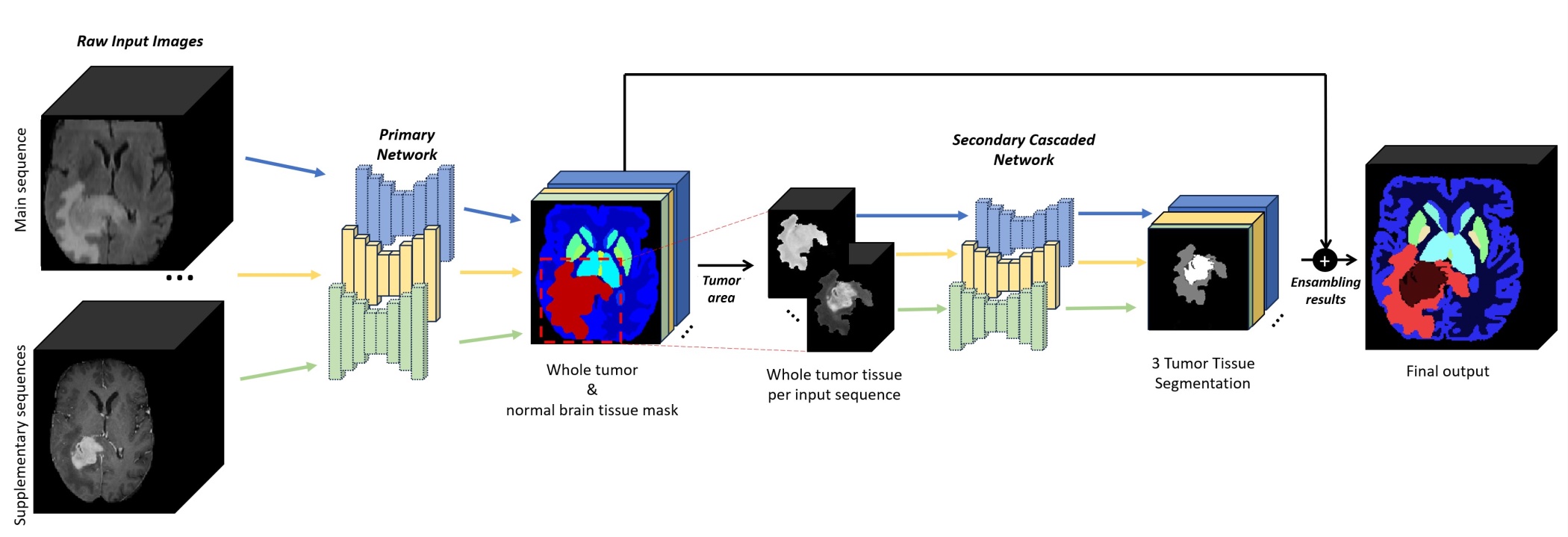
Figure: Cascaded architecture to produce the final output. For each input image, the first network segments the brain anatomy and potential tumors, while the second network subdivides tumors into 3 classes (Enhancing Tumor, Non-Enhancing Tumor, and Necrosis). Post-processing for refining tumor structure will be executed if multi-modal is available.
Installation
For running mri_TumorSynth follow the instructions in this webpage to create a virtual python environment with all the required dependencies and download the model files required for inference (see Links section).
Prerequisites
1. Install conda and locate the path to the activation file, this will be named 'conda.sh'
Instructions for installing conda can be found on their site here
- Once conda is installed, you can locate the path to your 'conda.sh' script by running one of the following commands:
bash # Linux cat ~/.bashrc | grep conda.shzsh # MacOS (if conda was initialized for zsh) cat ~/.zshrc | grep conda.sh
2. Download the model and the weights
The whole tumor model file is available for download from the TumorSynth Whole Tumor Weights Download Page.
- Navigate to the above link and click the 'Download/Arrow' button in the top left of the window
- Once the zip file containing the model is downloaded, unpack it. This can be done with the 'Archive Utility' on MacOS, or with the command below on either a Linux or MacOS system, after navigating to the directory that contains the zip file:
bash unzip TumourSynth_v1.0.zip
The above command will unpack the zip file, and create a directory named TumourSynth_v1.0 containing the nnUNet model dependencies. Rename this directory to Task002_Tumor.
bash mv TumourSynth_v1.0 Task002_Tumor
Note the location of this directory, as we will need to pass the location to the installation script. For the inner tumor segmentation, download the weights from this link TumorSynth Inner Tumor Weights Download Page and repeat two last steps (unzip) and rename to Task003_InnerTumor.
Installation script
Once the prerequisites have been met, you can run create_nnUNet_v1.7_env.sh to setup the nnUNet environment. The script will create a conda env with the tested version of python and pytorch, clone and install the version of nnUNet used to train the model, create the directory structure to hold the data and model files for nnUNet, generate a file to source with the env vars required by nnUNet to find the data.
The following args will need to be passed to the create_nnUNet_v1.7_env.sh script to correctly configure the nnUNet environment:
- The location of your 'conda.sh' script
- The location of the model file that you have downloaded an unpacked
- The name you want to give the conda env
- The location of your nnUNet installation
Example command:
bash ./create_nnUNet_v1.7_env.sh \ -e linux/etc/profile.d/conda.sh \ # path to conda.sh script -m Downloads/Task002_Tumor \ # path to unpacked model -n nnUNet_v1.7 \ # name to give the conda env -d /home/user_example/nnUNet # root of the nnUNet installation -c # if passed, install cuda
NOTE: Do not pass the -c flag on a Mac, cuda is not available for MacOS.
When the script finishes running (which could take a bit of time), you will have a new conda environment with the name passed to the -n flag. A file will also be created in your current directory ($PWD), named nnUNet_v1.7_path.sh that contains the environment variables that need to be defined for nnUNet to function properly. Source that file to set the required environment variables.
To have the nnUNet env vars set automatically when the conda environment is activated, move a copy of the nnUNet_v1.7_path.sh into the 'activate.d' directory in your conda environment.
With the newly created conda env active, run the following commands to create the 'activate.d' directory for the conda environment and move a copy of the nnUNet_v1.7_path.sh file there:
bash mkdir -p $CONDA_PREFIX/etc/conda/activate.d cp nnUNet_v1.7_path.sh $CONDA_PREFIX/etc/conda/activate.d
For the second set of weights (Task003_InnerTumor), you can run the script twice or copy/move them to your equivalent /home/user_example/nnUNet/nnUNet_v1.7/nnUNet_trained_models/nnUNet/3d_fullres directory.
Download main script
Once all the required weights and tools are installed, you can download the main script to run mri_tumorsynth from this GitHub repository: mri_TumorSynth main script
Key links
Conda installation: Conda website
Whole tumor model and weights: TumourSynth_v1.0.zip
Inner tumor model and weights: Task003_InnerTumor.zip
Installation script: create_nnUNet_v1.7_env.sh
mri_TumorSynth main script: mri_TumorSynth
Synopsis
mri_TumorSynth --i inputvol_or_list_inputvols --o outputvol [--wholetumor --innertumor --cpu --threads <threads>]
Arguments
Required Arguments
--i inputvol |
Input volume or list of input volumes separate by comma |
Path to primary MR scan (required; T1CE recommended for best results). Supports NIfTI (.nii/.nii.gz) format. It is also possible to provide multiple sequences separated by commas. All sequences must be registered and skull-stripped. |
--o outputvol |
Output volume |
Path to save segmentation mask (same format as input). |
Optional Arguments
--wholetumor |
Whole-tumor + healthy tissue mode |
Outputs combined mask of healthy brain tissue and whole tumor (includes necrosis, enhancing tumor, non-enhancing tumor). Input must be skull-stripped and registered to SRI-24 template. |
--innertumor |
Inner tumor substructure mode |
Outputs BraTS-compliant subclasses: Enhancing Tumor, Non-Enhancing Tumor, and Necrosis. Input must be a tumor ROI image (prepare by multiplying raw scan with --wholetumor output mask). |
--cpu |
Force CPU processing |
Bypasses GPU detection and runs on CPU (slower but compatible with systems without GPU). |
--threads <threads> |
Number of CPU threads |
Number of cores to use for CPU processing (default: 1; max: number of system cores). |
Usage
mri_TumorSynth supports three primary usage scenarios, with flexibility for single or multi-sequence inputs:
Basic Usage (Single Sequence)
Whole-tumor + healthy tissue segmentation (primary use case):
mri_TumorSynth --i t1ce_skullstripped.nii.gz --o tumor_whole_healthy_mask.nii.gz --wholetumor
Inner tumor substructure segmentation (requires tumor ROI input):
mri_TumorSynth --i t1ce_tumor_roi.nii.gz --o tumor_inner_substructures.nii.gz --innertumor
Multi-Sequence Usage (Improved Accuracy)
Combine T1CE, T2, and FLAIR for better segmentation of heterogeneous tumors:
mri_TumorSynth --i t1ce.nii.gz,t2.nii.gz,flair.nii.gz --o tumor_multi_seq_mask.mgz --wholetumor
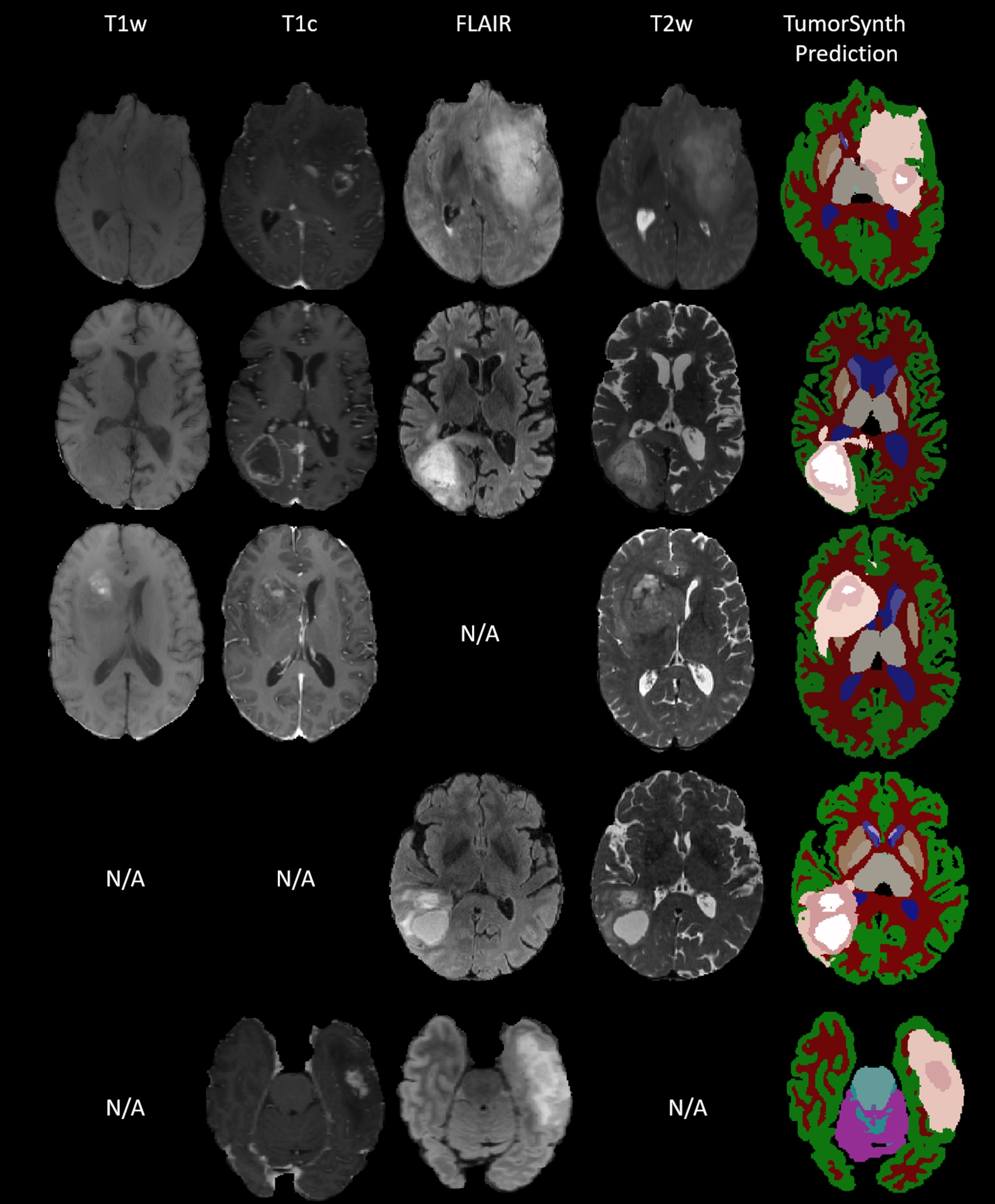
Figure: TumorSynth’s performance on example cases. Post-processing for refining tumour structure will be executed if multi-modal is available. N/A stands for missing this modality when doing the inference.
Examples
Example 1: Whole-Tumor + Healthy Tissue Segmentation (Single Sequence)
mri_TumorSynth --i t1ce_skullstripped_SRI24.nii.gz --o t1ce_whole_tumor_healthy_mask.nii.gz --wholetumor
Input: Skull-stripped T1CE scan registered to SRI-24 template.
Output: Mask containing healthy brain tissue (e.g., white matter, gray matter, CSF) and whole tumor (necrosis + enhancing + non-enhancing tumor).
Example 2: Inner Tumor Substructure Segmentation
First, prepare the tumor ROI input (using --wholetumor output):
fslmaths wholetumorsegmentation.nii.gz -thr 17.5 -uthr 18.5 -bin -mul t1ce.nii.gz t1ce_tumor_roi.nii.gz
Then run inner tumor segmentation:
mri_TumorSynth --i t1ce_tumor_roi.nii.gz --o t1ce_inner_tumor_substructures.nii.gz --innertumor
Output: BraTS-compliant labels for Necrosis, Non-Enhancing Tumor (NET), and Enhancing Tumor (ET).
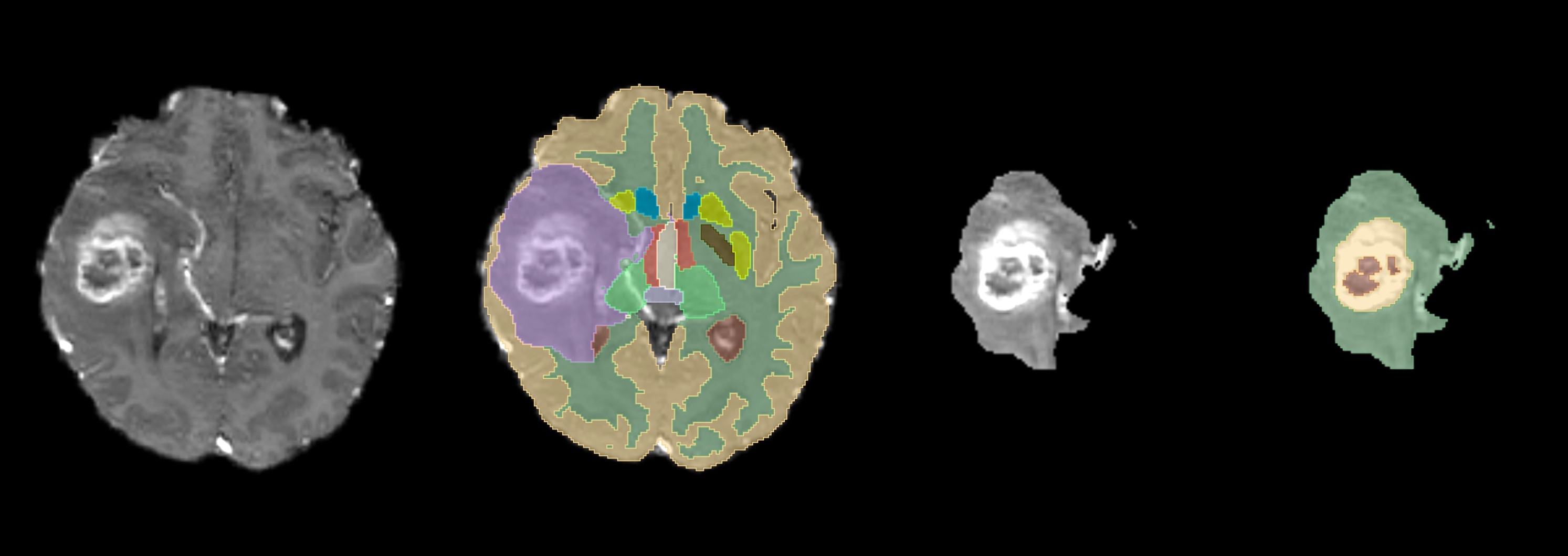
Figure: From left to right, all the steps for getting the inner tumor segmentation. First the input T1CE, second the whole tumor segmentation, third the isolated tumor area from the T1CE, and finally, the inner tumor segmentations following BraTS-compliant labels.
Example 3: Multi-Sequence Segmentation
mri_TumorSynth --i t1ce.nii.gz,t2.nii.gz,flair.nii.gz,t1.nii.gz,adc.nii.gz --o tumor_multi_seq_mask.nii.gz --wholetumor --threads 4
Outputs: Segmentation mask with all healthy tissues and whole tumor.
List of Segmented Structures
Segmentation labels follow FreeSurfer’s anatomical classification for healthy tissue and BraTS criteria for tumor substructures.
#No. |
Label Name: |
0 |
Unknown |
1 |
Cerebral-White-Matter |
2 |
Cerebral-Cortex |
3 |
Lateral-Ventricle |
4 |
Inferior-Lateral-Ventricle |
5 |
Cerebellum-White-Matter |
6 |
Cerebellum-Cortex |
7 |
Thalamus |
8 |
Caudate |
9 |
Putamen |
10 |
Pallidum |
11 |
3rd-Ventricle |
12 |
4th-Ventricle |
13 |
Brain-Stem |
14 |
Hippocampus |
15 |
Amygdala |
16 |
Accumbens-Area |
17 |
Ventral-DC |
18 |
Whole tumor |
Table: Label values and corresponding structures for --wholetumor mode.
#No. |
Label Name: |
0 |
Unknown |
1 |
Non-enhancing |
2 |
Enhancing |
3 |
Necrosis |
Table: Label values and corresponding structures for --innertumor mode.
Frequently Asked Questions (FAQ)
Do I need to preprocess input scans?
For --wholetumor mode: Inputs must be skull-stripped and registered to the SRI-24 template (FreeSurfer’s recon-all or mri_robust_register can be used). No additional preprocessing (e.g., bias field correction, intensity normalization) is required.
For --innertumor mode: Inputs must be tumor ROI images (prepared by masking the raw scan with --wholetumor output).
What MR sequences are supported?
T1CE is the recommended primary sequence. T2 and FLAIR can be added as secondary/tertiary inputs for improved accuracy, but other sequences can be provided. The tool is agnostic to the input sequence type, and fuses the information from all the input sequence segmentations for obtaining the final segmentation.
Why is my segmentation misaligned with the input scan?
Misalignment occurs if the input is not registered to the SRI-24 template (required for --wholetumor mode). Re-register the input using mri_robust_register and re-run the tool. For non-FreeSurfer viewers, save the registered input with --resample (compatible with FreeSurfer v7.4.1+).
How can I speed up processing?
Use a GPU (default if compatible; 5-10x faster than CPU). For CPU processing: Increase the --threads flag (e.g., --threads 8 for an 8-core machine).
Avoid multi-sequence input if speed is prioritized (single T1CE is fastest).
What file formats are supported?
Inputs and outputs: NIfTI (.nii/.nii.gz) format.
See Also
mri_synthseg, recon-all, mri_robust_register, mri_volstats
References
SynthSeg: Segmentation of brain MRI scans of any contrast and resolution without retraining. B Billot, et al. Medical Image Analysis, 83, 102789 (2023).
BraTS 2021 Challenge: Brain Tumor Segmentation Challenge.
Reporting Bugs
Report bugs to analysis-bugs@nmr.mgh.harvard.edu with the following information: FreeSurfer version (run freesurfer --version). Exact command used to run mri_TumorSynth. Error message (copy-paste full output). Input file details (sequence type, format, resolution).
Author/s
JaneSmith and JiamingWu and FerranPrados