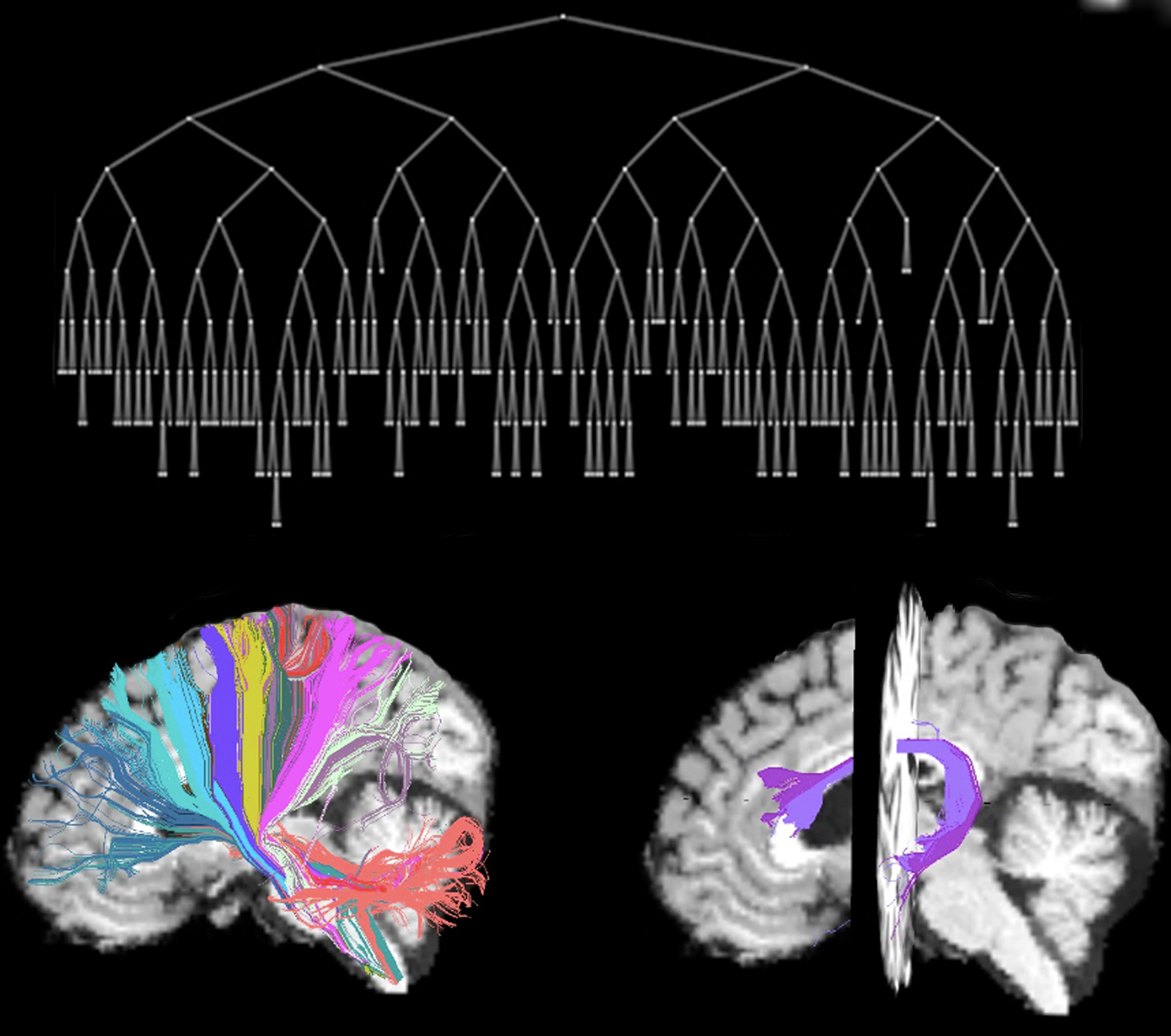
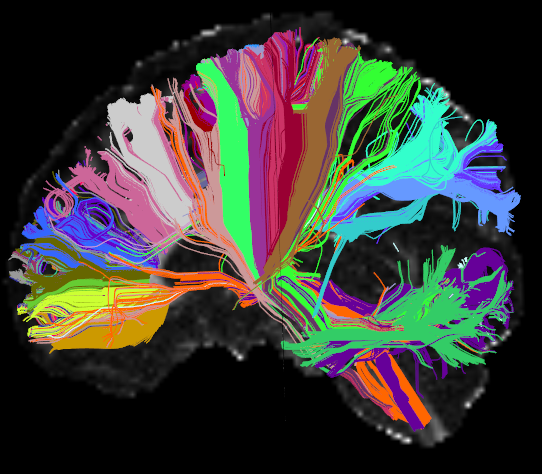
AnatomiCuts is an unsupervised hierarchical clustering that uses an anatomical similarity metric to cluster together white matter streamline with similar neighboring anatomical structures.
Contents
AnatomiCuts stream
Enviroment variables
First you will need to set your SUBJECTS_DIR. This directoy should contain your recon outputs (mri folder). It is also necesary the dmri_preproc output (dmri folder). If this command was run on another directory, you can specify it with the DMRI_DIR variable. Additionally, you can specify a different output directory by setting ODMRI_DIR. By default, everything will point to your SUBJECTS_DIR.
setenv SUBJECTS_DIR "dir with recon folders (mri)"
Optionally, you can have dmri_preproc and dmri.ac in different folders but setting the following variables:
setenv DMRI_DIR "dir with mri_preproc outputs (dmri)" setenv ODMRI_DIR "output dir for dmri.ac (anatomicuts)"
AC-All
The following command will run every preprocessing step and AnatomiCuts. - MODEl can be DTI or DKI. If DKI, microstructural maps will be obtain for DKI and also DTI. DKI requires more than one bvalue. - Lenght is a parameter to filter short streamlines. Depending on age or population, this number could range from 35mm to 55mm for filtering spurious but also some u-shape fibers.
dmri_ac.sh runAC <<SUBJECT_ID>> <<MODEL>> <<LENGHT>>
example:
dmri_ac.sh runAll bert DKI 45
Additionally, if you are in the center, you can run it on the cluster for all subjects by adding modifying the command as follows:
dmri_ac.sh forAll runAC <<MODEL>> <<LENGHT>> "pbsubmit -n 1 -c "
where "pbsubmit -n 1 -c " is the command that is executed in the cluster. If you don't pass any string, the command will be run for every subject sequentially.
AnatomiCuts executable
If you only want to run AnatomiCuts, maybe with another set of streamlines that you have. For this, it is required to have a parcellation and a streamline dataset on the same space.
Inputs
- streamlines file (*.trk)
- segmentation image, ideally, including cortical and subcortical parcellation with white matter segmentation based on neighboring regions (wmparc or wm2009parc).
It is important to ensure that the white matter streamlines and the segmentation are in the same space.
Filtering streamlines
Sometimes deterministic tractography can generate spurious streamlines outside the brain, or very short cut off streamlines. To exclude them we can use streamlineFilter. It is recommended to remove the short streamlines that can prematurely end during tractography. It is also possible to remove the ushape streamlines.
Running AnatomiCuts
AnatomiCuts -s segmentation.nii.gz -f streamlines.vtk -labels -o outputFolder <options>
Where
-s segmentation file such as wm2009parc.nii.gz
-f streamline file in trk format.
-c number of clusters. The default is 200.
-n number of points to be sampled per streamline. The default is 10.
-e number of streamlines to be used for the eigendecomposition of normalized cuts algorithm. The default is 500. Using less number of streamlines will fasten substantially the algorithm at the expense of accuracy. For a quick test 50 could be ok, but in practice, less than 300 is not recommended.
-d directional neighbors to be used, the default is "a" all with 26 directions, diagonal "d" contains 14 directions and straight "s" only 6 directions.
-o output folder
Finding corresponding clusters across subjects
The Hungarian algorithm allows to find one-to-one correspondences and it's implementation for AnatomiCuts at multiple levels of the tree hierarchy can be found in AnatomiCuts_correspondences.
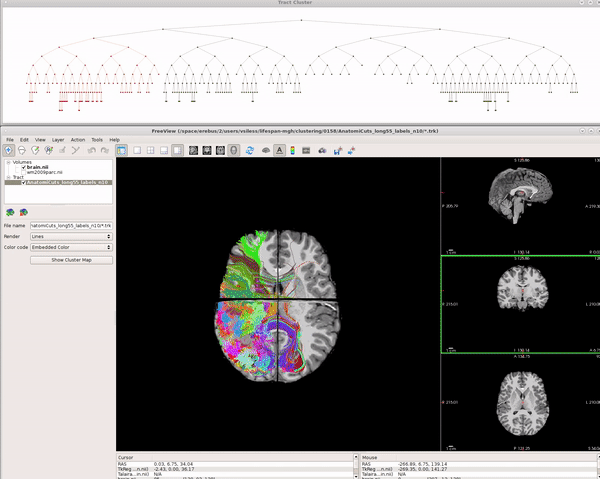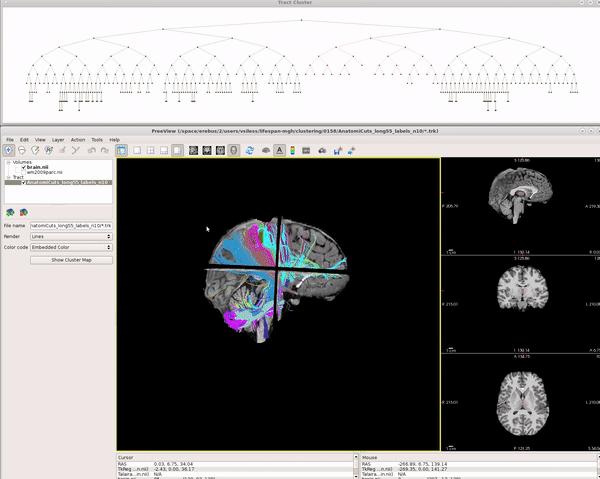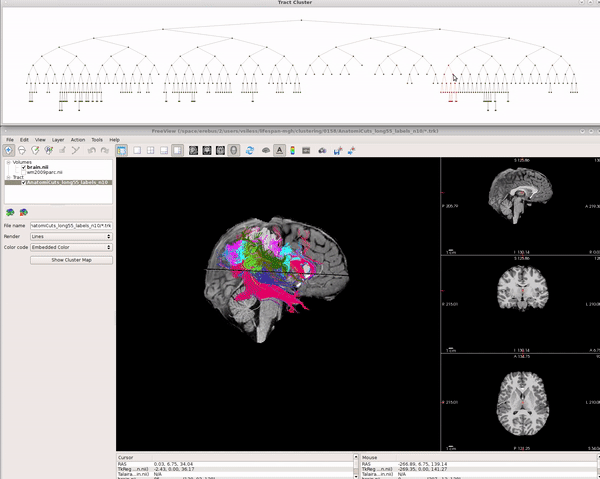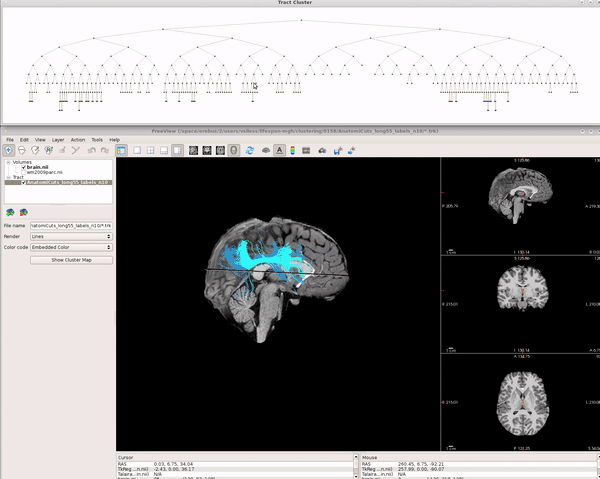
Visualizing AnatomiCuts in Freeview
To load the clusters obtained with AnatomiCuts go to "File -> Load Tract Cluster" and select the AnatomiCuts output folder.
References
AnatomiCuts: Hierarchical clustering of tractography streamlines based on anatomical similarity. Siless V., Chang K., Fischl B., Yendiki A.. NeuroImage 2018.
V. Siless, J. Y. Davidow, J. Nielsen, Q. Fan, T. Hedden, M. Hollinshead, C. V. Bustamante, M. K. Drews, K. R. A. Van Dijk, M.A. Sheridan, R. L. Buckner, B. Fischl, L. Somerville, and A. Yendiki. 2017. “Registration-free analysis of diffusion MRI tractography data across subjects through the human lifespan.”