LCN Cortical Lobes Dataset
The LCN-CortLobes dataset is a collection of manually-inspected cortical lobe labels created from FreeSurfer-processing the T1w MR images of 35 subjects, ages 64 - 96, originating from the ADNI and OASIS datasets. The set is intended to supplement the Mindboggle-101 dataset of cortical labels Klien and Tourville, 2012, which lack image data of subjects over the age of 61, and lack image data acquired on GE scanners. Unlike the Mindboggle set, only the cortical lobes (frontal, parietal, temporal and occipital) have been manually inspected. The lobe labels are composed from the freesurfer-generated cortical regions, following the DKT labeling protocol, as described in the appendix of the Klein and Tourville paper. The DKT cortical regions in the LCN-CortLobes dataset have not been manually inspected.
The set is freely available for download (see Data Download below). All image-processing and visual inspection efforts were conducted at the Laboratory for Computational Neuroimaging (LCN).
Methods
Data
To address two shortcomings in the Mindboggle dataset, images from publicly available datasets were identified following this criteria:
- subjects over the age of 61
- subjects scanned on GE MRI scanners
34 subjects were identified in the ADNI dataset, and 1 subject from the OASIS. The selected T1-weighted images were free of image artifacts. The demographic and scanner information is shown in the table below (included in the dataset distribution). Note that several subjects were diagnosed with Alzheimer's Disease (AD), mild cognitive impairment (MCI), and late-stage MCI (LMCI). The remaining were cognitively normal (CN).
subject_id |
age |
sex |
manufacturer |
field_strength |
diagnosis |
source |
002_S_0295_13722 |
84.9041 |
M |
GE |
1.5 |
CN |
ADNI |
002_S_0413_14437 |
76.3863 |
F |
GE |
1.5 |
CN |
ADNI |
002_S_0685_18211 |
89.737 |
F |
GE |
1.5 |
CN |
ADNI |
002_S_0685_MR1 |
93 |
F |
GE |
1.5 |
CN |
ADNI |
002_S_0782_20519 |
81.7068 |
M |
GE |
1.5 |
MCI |
ADNI |
002_S_0816_49010 |
71.4849 |
M |
GE |
1.5 |
AD |
ADNI |
002_S_0954_108600 |
71.0795 |
F |
GE |
1.5 |
MCI |
ADNI |
003_S_0907_MR1 |
95 |
F |
Siemens |
1.5 |
CN |
ADNI |
003_S_1074_MR1 |
91 |
F |
Siemens |
1.5 |
MCI |
ADNI |
005_S_0221_11604 |
67.5315 |
M |
GE |
1.5 |
AD |
ADNI |
005_S_0222_11299 |
85.9918 |
M |
GE |
1.5 |
MCI |
ADNI |
005_S_0602_66117 |
72.0685 |
M |
GE |
3 |
CN |
ADNI |
005_S_1341_43188 |
71.6521 |
F |
GE |
1.5 |
AD |
ADNI |
006_S_0322_17197 |
65.6658 |
M |
GE |
1.5 |
MCI |
ADNI |
006_S_6441_MR1 |
89 |
M |
Philips |
3 |
MCI |
ADNI |
007_S_0101_9602 |
73.6712 |
M |
GE |
1.5 |
LMCI |
ADNI |
007_S_0128_10007 |
64.1699 |
F |
GE |
1.5 |
MCI |
ADNI |
009_S_0842_24339 |
73.7315 |
M |
GE |
1.5 |
CN |
ADNI |
009_S_0862_25128 |
73.5041 |
F |
GE |
1.5 |
CN |
ADNI |
009_S_1030_28514 |
67.5507 |
M |
GE |
1.5 |
LMCI |
ADNI |
029_S_1073_30359 |
65.7699 |
F |
GE |
1.5 |
MCI |
ADNI |
032_S_6293_MR1 |
88 |
F |
Siemens |
3 |
CN |
ADNI |
094_S_1293_98699 |
78.2466 |
M |
GE |
3 |
MCI |
ADNI |
098_S_0172_11398 |
70.6904 |
F |
GE |
1.5 |
CN |
ADNI |
126_S_0606_37834 |
69.1397 |
F |
GE |
3 |
AD |
ADNI |
126_S_0784_25775 |
76.0055 |
F |
GE |
1.5 |
AD |
ADNI |
127_S_0259_11888 |
70.663 |
M |
GE |
1.5 |
CN |
ADNI |
127_S_0260_28530 |
79.3945 |
F |
GE |
3 |
CN |
ADNI |
127_S_0393_30421 |
86.8466 |
F |
GE |
3 |
MCI |
ADNI |
127_S_0754_23787 |
67.7178 |
F |
GE |
1.5 |
AD |
ADNI |
127_S_0844_29229 |
85.4603 |
F |
GE |
3 |
AD |
ADNI |
131_S_0384_14252 |
79.9616 |
M |
GE |
3 |
LMCI |
ADNI |
131_S_1389_78461 |
76.8192 |
M |
GE |
3 |
LMCI |
ADNI |
133_S_0525_16133 |
70.1425 |
F |
GE |
1.5 |
CN |
ADNI |
OAS1_0278_MR1 |
96 |
F |
Siemens |
1.5 |
HC |
OASIS |
Processing
The standard processing stream ("recon-all -all") of FreeSurfer v7.1.0 was used to create subcortical and cortical segmentations and parcellations. All images completed processing without errors. FreeSurfer automatically creates cortical parcellations following the DKT labeling protocol (described in the appendix of the Klein and Tourville paper). The FreeSurfer utility "mri_annotation2label --lobesStrict ?h.DKTlobes.annot --annotation ?h.aparc.DKTatlas.annot" was used to create the cortical lobe surface label files (l/rh.DKTlobes.annot). "mri_annotation2label --lobesStrict" implements the region-to-lobe mapping described in the Appendix of this publication. See also CorticalParcellation.
Inspection
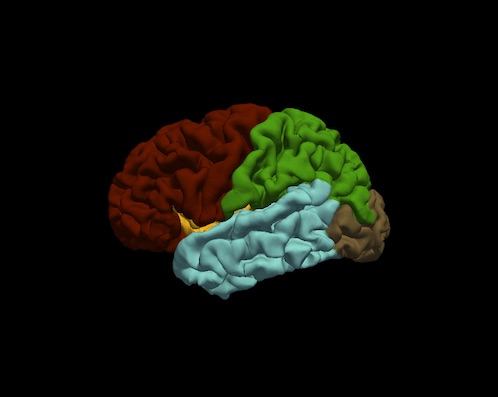
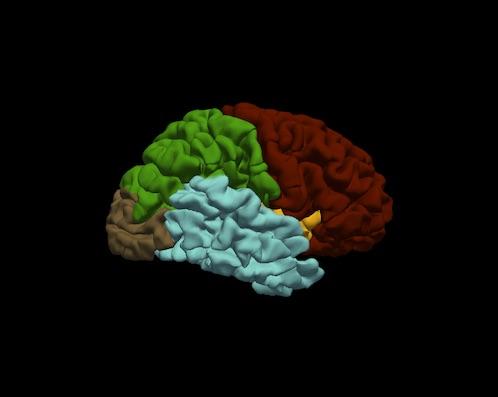
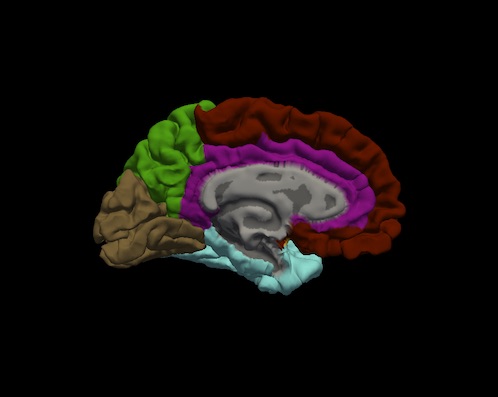
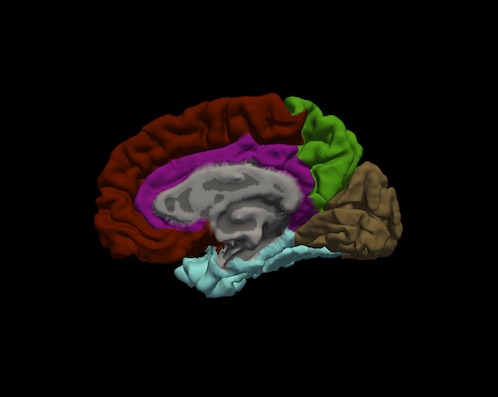
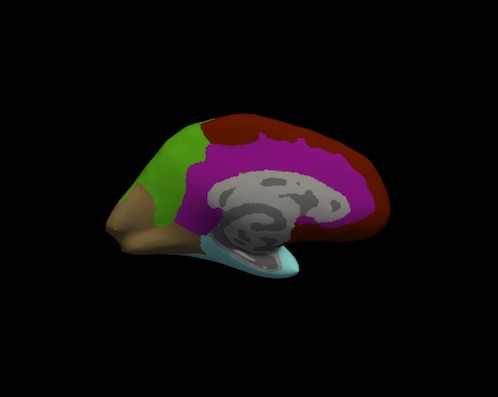
Bruce Fischl (BF), Ph.D., Professor in Radiology at Harvard Medical School, Neuroscientist at Massachusetts General Hospital, conducted a visual inspection of image snapshots of the cortical lobe labels. Inspection involved verifying the cortical lobe boundaries created by the process described in Klein and Tourville. For each subject, and for each hemisphere, a snapshot of the lateral and medial surfaces, both 'pial' and 'inflated' surfaces, were generated by the FreeSurfer utility "freeview". An example of the set of snapshots generated for each subject is shown below. The frontal lobe is red, the temporal lobe is blue, the parietal lobe is green and the occipital lobe is brown. Note: the insula is yellow and cingulate is purple.
|
|
|
|
|
|
|
|
Results
The visual inspections conducted by BF of the cortical lobe snapshots found no discernable problems with the lobe labels (as stored in the files l/r.DKTlobes.annot) in the 35 subjects.
Data Download
Click this link to download the tarball: LCN-CortLobes.tgz
Uncompress it with the command 'tar zxvf LCN-CortLobes.tgz'.
The set includes the following files:
- demographics.tsv - spreadsheet of info on the set's 35 subjects:
- subject_id
- age (64 - 96)
- sex (M or F)
- manufacturer (GE, Siemens or Philips)
- field_strength (1.5 or 3)
- diagnosis (CN = cognitively normal, MCI = mild cognitive impairment, LMCI = late-stage MCI, AD = Alzheimer's Disease)
- source (ADNI or OASIS)
<subject_id>/mri - folder containing:
- orig.mgz - the T1w input
- aparc.DKTatlas+aseg.mgz - the labeled volume of DKT cortical regions and subcortical structure segmentations (note: not an inspected file)
- aparc.DKTlobes+aseg.mgz - the labeled volume of cortical lobes and subcortical structure segmentations (note: not an inspected file)
<subject_id>/label - folder containing:
- ?h.aparc.DKTatlas.annot - the surface labels for the DKT cortical regions (note: not manually inspected)
?h.DKTlobes.annot - the surface labels for the cortical lobes, created from mri_annotation2label --lobesStrict, manually inspected
- lobesQA - folder containing snapshots taken of the cortical surface showing the lobes, used in the manual inspection process
Conflict of Interest Statement
BF has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. BF's interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Contributions
Nick Schmansky (NS) of CorticoMetrics LLC originated the idea for the creation of this dataset, as a means to fulfill FDA 510(K) software validation requirements. NS identified and collected the image set and provided the data to engineers at the Laboratory for Computational Neuroimaging (BF is the Lab Director), where the data was processed by FreeSurfer and inspected by BF. NS created this wiki page, and BF contributed to its contents.