Back to list of all tutorials | Back to course page | Previous | Next (Directory Structure)
Contents
1. FSFAST Tutorial Data Description
The functional data were collected as part of the Functional Biomedical Research Network (fBIRN, www.nbirn.net).
- Working-memory paradigm with distractors
- 18 subjects
- Each subject has 1 run (except sess01 which has 4 runs)
- Collected at MGH Bay 4 (3T Siemens)
FreeSurfer anatomical analyses
2. Functional Paradigm
The paradigm was designed to study the effects of emotional stimuli on the ability to recall items stored in working memory.
- Blocked design
- Each block consisted of 3 phases
- Encode (16 sec) - 8 stick figures to remember (no response)
- Distractor (16 sec) - 8 distractor images (response whether there is a face in the image)
- Emotional - Distractors are emotionally disturbing
- Neutral - Distractors are emotionally neutral
- Probe (16 sec) - 8 pairs of stick figures. Subject responds as to which of the pair was in the original Encode.
- Between each block was a 16 sec scrambled image used as baseline.
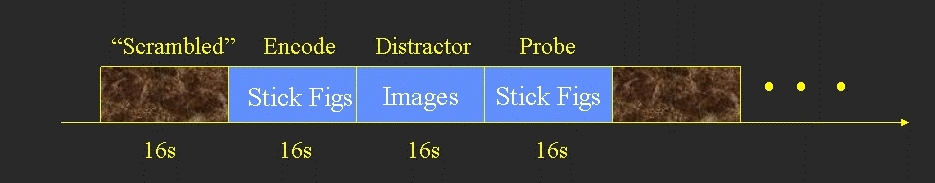
The above yields 5 conditions:
- Encode
- Emotional Distractor
- Neutral Distractor
- Probe following Emotional Distractor
- Probe following Neutral Distractor
The scrambled image will be modeled as a baseline, not as a condition.
3. Functional Data
- Original data: each subject had 8 runs
- This data: each subject has 1 run (except for sess01 who has 4)
- Each run lasts 142 time points
- TR = 2 sec.
- There is one run of rest data for 13 subjects
- There is a B0 map for each subject
4. Anatomical Data
FreeSurfer analysis has been run for all 18 subjects
5. Getting the Data (not necessary for the Boston FreeSurfer Course)
You can get the analyzed functional data (10G) from:
wget ftp://surfer.nmr.mgh.harvard.edu/pub/data/fsfast-functional.tar.gz
You can get the structural data (5G) from:
wget ftp://surfer.nmr.mgh.harvard.edu/pub/data/fsfast-tutorial.subjects.tar.gz
6. Organizing the Tutorial (not necessary for the Boston FreeSurfer Course)
cd to a place on your network where you have enough space to unpack the tutorial data.
cd /place/with/space
Untar the data
tar xvfz fsfast-tutorial.tar.gz tar xvfz fsfast-tutorial.subjects.tar.gz
You will need to set the TUTORIAL_DATA environment variable. In bash, run:
export TUTORIAL_DATA=/place/with/space
In tcsh or csh
setenv TUTORIAL_DATA /place/with/space
You will also need to link the FreeSurfer anatomical subjeccts (data in fsfast-tutorial.subjects) into your $SUBJECTS_DIR.
You should set the FSFAST output format to be compressed NIFTI (nii.gz). In bash, run:
export FSF_OUTPUT_FORMAT=nii.gz
In tcsh or csh, run:
setenv FSF_OUTPUT_FORMAT nii.gz
Back to list of all tutorials | Back to course page | Previous | Next (Directory Structure)
