Longitudinal Processing - Tutorial
This page will take you through the steps of processing your longitudinal data: first with running the cross sectionals and creating the unbiased within-subject base and longitudinals. Then we will take a look at the results, learn how to do some post-processing and about editing the data and rerunning the different streams with the new edits. You can read more about the longitudinal stream at LongitudinalProcessing and about edits at LongitudinalEdits.
Preparations
Note: The tutorial data has been processed with Freesurfer 5.1. The commands, as described below, to view or post-process the data will work with newer versions of Freesurfer. However, if you use a newer version of Freesurfer to actually process the data on your own (recon-all) you may obtain different results as depicted below.
If You're at an Organized Course
If you are taking one of the formally organized courses, everything has been set up for you on the provided laptop. The only thing you will need to do is run the following commands in every new terminal window (aka shell) you open throughout this tutorial. Copy and paste the commands below to get started:
setenv SUBJECTS_DIR $TUTORIAL_DATA/long-tutorial cd $SUBJECTS_DIR
To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.
These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into this directory. You can now skip ahead to the tutorial (below the gray line).
If You're not at an Organized Course
If you are NOT taking one of the formally organized courses, then to follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. These are the commands that you need to run before getting started:
tcsh source your_freesurfer_dir/SetUpFreeSurfer.csh setenv SUBJECTS_DIR your_tutorial_data/long-tutorial cd $SUBJECTS_DIR
Notice the command to open tcsh. If you are already running the tcsh command shell, then the 'tcsh' command is not necessary. If you are not using the tutorial data you should set your SUBJECTS_DIR to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located.
Creating Longitudinal Data
Warning: Running recon-all comands can take a significant amount of time (up to 24h). Therefore do not run the commands in this section if you are participating in one of our organized courses. All the data has already been processed for you for the course!
Processing your data currently consists of three steps:
First, run all your data independently (we also say cross sectionally) using "recon-all -all" (i.e. all time points for all subjects):
recon-all -subjid <tpNid> -all
Second, create the within-subject templates (also called "base", not to be confused with baseline, i.e. the initial scan!) from the tpNs for each subject. Here you can choose a name for the templateID, e.g. 'bert' or 'bert_base' (should not be identical to one of the time point ids):
recon-all -base <templateID> -tp <tp1id> -tp <tp2id> ... -all
Finally, create the longitudinals using the subject template and tpNs. Repeat the following steps for all tpNs. The resulting directories will be in the format of tp1id.long.templateID
Important Note! If you are re-processing existing longitudinal data processed by v5.0 or v4.5, and you are using v5.1 or newer, be sure to add the -clean flag to your recon-all string, as you will want to overwrite the brainmask.mgz and other files, since the base and long subject space is in a different space than in pre v5.1 runs. This is true for the -long stage as well.
recon-all -long <tpNid> <templateID> -all
Let's step through a full example. For a subject with two time points OAS2_0001_MR1 and OAS2_0001_MR2 you would run the independent runs first by typing (don't do it, it has already been done for you) in two separate terminals or on different computers:
recon-all -subjid OAS2_0001_MR1 -all recon-all -subjid OAS2_0001_MR2 -all
If you work with your own data, you'd have to import the dicoms first recon-all -i path_to_your_dcm1 ... -subjid your_subject_name. In this command you can pass several within-session scans by specifying multiple -i flags instead of the "...". You then run the above recon-all comands to process the imported data. We call these runs the cross sectionals (or cross runs) because the two time points are processed completely independently as if they were from different subjects.
Once the norm.mgz is available on both time points, you can create the unbiased subject template/base. We decide to name it OAS2_0001 :
recon-all -base OAS2_0001 -tp OAS2_0001_MR1 -tp OAS2_0001_MR2 -all
This will create the within-subject template (we will refer to these type of runs as base) and run it through recon-all. It will take approximately the same time as a regular recon-all run, a little more because of the initial averaging of time points. A directory OAS2_0001 will be created.
Finally once the base and the two cross sectionally processed time points are fully completed, you can run the longitudinal runs (again in two different terminals or different computers) by typing:
recon-all -long OAS2_0001_MR1 OAS2_0001 -all recon-all -long OAS2_0001_MR2 OAS2_0001 -all
Warning: Running recon-all comands can take a significant amount of time (up to 24h). Therefore do not run the commands in the prior sections if you are participating in an organized course. All the data has already been processed for you for the course! Of course you will need to run those recon-all commands on your own data.
These runs create the directories OAS2_0001_MR1.long.OAS2_0001 and OAS2_0001_MR2.long.OAS2_0001 containing the final results. These are complete subjects directories and we will use them for any post-processing or analysis as the results are more sensitive and repeatable than the independent cross runs. These longitudinal processes run much faster than the cross and base above. We call them the longitudinal or simply long runs, because they make use of common information taken from the template.
Inspecting Longitudinal Data
Once your results are there (and they are in this tutorial), you can take a look at different things. First let's check the base. Open the norm.mgz with freeview (a nice viewer):
freeview -v OAS2_0001/mri/norm.mgz \
-f OAS2_0001/surf/lh.pial:edgecolor=red \
OAS2_0001/surf/rh.pial:edgecolor=red \
OAS2_0001/surf/lh.white:edgecolor=blue \
OAS2_0001/surf/rh.white:edgecolor=blue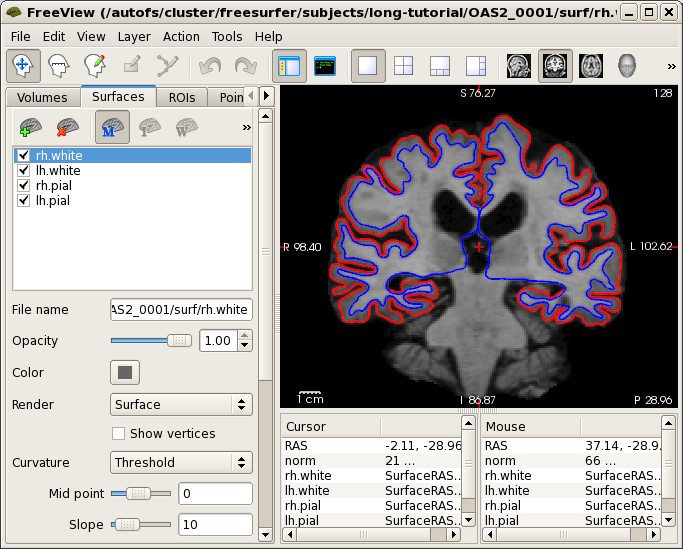
This will show you a synthesized image, basically the average anatomy of this subject across time overlayed with the pial and white surface. If the across-time registration failed you would see a blurry image or ghosts (usually never happens, but if it does, report it). You can also inspect the surfaces on the average anatomy. This will be important later as the surfaces are transferred into the longitudinal runs and therefore should be accurate in the base.
Now it is time to look at the longitudinal results. Starting with Freesurfer 5.1, the base and the long runs are all in the same voxel space, therefore images will be registered and can be directly compared if opened on top of each other. Close any open freeview to reduce memory usage and run the following command:
freeview -v OAS2_0001_MR1.long.OAS2_0001/mri/norm.mgz \
OAS2_0001_MR2.long.OAS2_0001/mri/norm.mgz \
-f OAS2_0001_MR1.long.OAS2_0001/surf/lh.pial:edgecolor=red \
OAS2_0001_MR1.long.OAS2_0001/surf/lh.white:edgecolor=blue \
OAS2_0001_MR2.long.OAS2_0001/surf/lh.pial:edgecolor=255,128,128 \
OAS2_0001_MR2.long.OAS2_0001/surf/lh.white:edgecolor=lightblue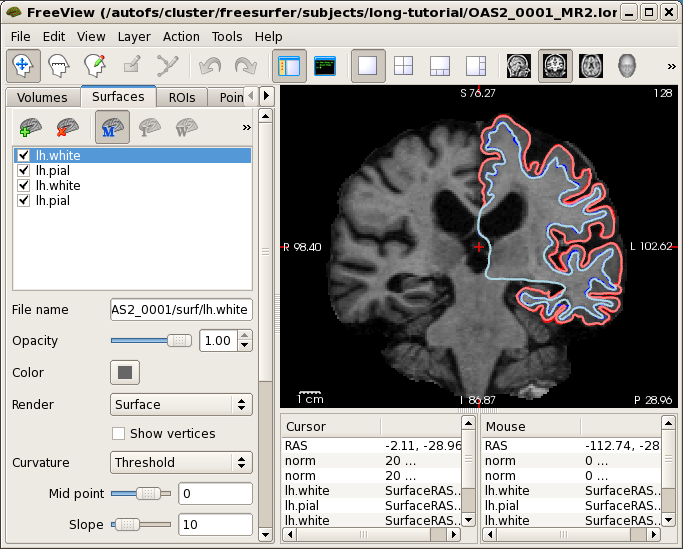
This might take a while to load (check the progress bar). We only open the surfaces in the left hemisphere to save time (and memory). You will see the normalized skull stripped images (norm.mgz) of both time points on top of each other (the second time point is the top-most layer and so the only one visible at the moment). You also see the white and pial surface for each time point, drawn on top of each other (we only show the left hemisphere here to reduce memory usage for older computers). Time point 1 uses red and blue while time point 2 uses pink and light blue for the surface colors. The surfaces do not change much across time. You can also compare the images directly. For this, uncheck the 4 check boxes in front of all the surfaces and click on the 'Volumes' tab which shows the two timepoint's norm.mgz files. Switch back and forth between the two images by pressing 'Alt-c'. Use 'Page Up' and 'Page Down' to scroll through the slices. You will mainly notice that the ventricles are getting larger in the second time point (and some non-linearities).
We use freeview as a viewer here, because tkmedit is limited to only two images and cannot load more than one set of surfaces. More info about freeview can be found in the FreeviewGuide.
Post-Processing Longitudinal Data
In order to analyze your longitudinal data, you have different options. You could, e.g., open the stats text files in each tpN.long.templateID/stats/ dir, containing statistics such as volume of subcortical structures or thickness averages for cortical regions. These statistics can be fed into any external statistical packages to run whatever analysis you are interested in, such as linear mixed effects models (often used in longitudinal analysis). Helpful commands to grab the data from all subject and create a single table are asegstats2table and aparcstats2table (since FS5.2 you can now pass a longitudinal qdec table to these scripts using the --qdec-long flag, see details below on the longitudinal qdec file format).
In the GLM and QDEC group analysis tutorials you learn how to run some simple statistical analyses on thickness maps (e.g. to find cortical regions with different thickness across groups). We won't do any statistical analysis in this tutorial, but we will discuss how to prepare and view your longitudinal data. Here we prepare for a simple statistical model for longitudinal data, consisting of 2 stages:
- we compute some measure for each subject, for example some measure of change such as mm/year thinning or percent change
- this measure can then be compared across groups (e.g. with QDEC) to detect disease or treatment effects.
Note: this 2-stage model is very simple. For more powerfull and complex modeling you should look at our tools for linear mixed effects.
Longitudinal QDEC Table
To get the longitudinal data ready for the 2 stage analysis you need to create a table (space separated as a text file) in the following format:
fsid |
fsid-base |
years |
... |
OAS2_0001_MR1 |
OAS2_0001 |
0 |
|
OAS2_0001_MR2 |
OAS2_0001 |
1.25 |
|
OAS2_0004_MR1 |
OAS2_0004 |
0 |
|
OAS2_0004_MR2 |
OAS2_0004 |
1.47 |
|
... |
|
|
|
where the first column is called fsid (containing each time point) and the second column is fsid-base containing the base name, to group time points within subject. You can have many more columns such as gender, age, group, etc. Make sure there is a column containing an accurate time variable (optimally measured in years if you are interested in yearly change) such as age or the duration of the study (time from inital scan). Here we use years to measure the time from baseline scan (=tp1). You can see in the table that the two subjects OAS2_0001 and OAS2_0004 each have two time points that are not equally spaced (approx 15 and 18 months apart). We have created this table for you in the qdec subdirectory for the tutorial data and can get started right away. You can look at the file by opening it in your favorite text/spreadsheet editor, e.g.:
gedit qdec/long.qdec.table.dat
you see, it is just a text file, so it looks a bit ugly. Better open it as a spreadsheet in Openoffice (select 'space' for separation when promted):
ooffice -calc qdec/long.qdec.table.dat
Preparing the Data - QCACHE
The following commands can be used to prepare the data (don't run it, it will take a while and has already been done for you):
long_mris_slopes --qdec ./qdec/long.qdec.table.dat --meas thickness --hemi lh --do-avg --do-rate --do-pc1 --do-spc --do-stack --do-label --time years --qcache fsaverage --sd $SUBJECTS_DIR
This will:
- (--qdec) read in the qdec table
- (--meas) take the thickness measure of each time point
- (--hemi) work on left hemisphere
- (--do-avg) compute the temporal average (thickness at midpoint of linear fit, here it is just the average)
- (--do-rate) compute the rate of change (thickening in mm/year)
- (--do-pc1) compute the percent change (with respect to time point 1)
- (--do-spc) compute a symmetrized percent change (with respect to the temporal average)
- (--do-stack) output a stacked thickness file for each subject (time series)
- (--do-label) intersect the cortex label to ensure we don't include non-cortex regions
- (--time) specify the column in the longqdec.table.dat that contains the time variable (here 'years')
- (--qcache) and automatically smooth everything on different levels and map it to fsaverage for a potential group analysis using qdec
- (--sd) specify the subject directory (needs to be passed explicitly since FS 5.2)
You then need to run the same command for the right hemisphere (--hemi rh). Note, if you split your table up into smaller tables containing only information for a specific subject each, you can run this on a cluster in parallel for each subject to speed things up (you can use cd qdec ; long_qdec_table --qdec ./long.qdec.table.dat --split fsid-base ; cd .. to split the table up into individual subjects inside the qdec directory).
Now before continuing, let's get an idea about what the above 4 measures mean in our setting (2 time points):
The temporal average is simply the average thickness: avg = 0.5 * (thick1 + thick2)
The rate of change is the difference per time unit, so rate = ( thick2 - thick1 ) / (time2 - time1), here thickening in mm/year, we expect it to be negative in most regions (because of noise not necessarily for a single subject, but on average).
The percent change (pc1) is the rate with respect to the thickness at the first time point: pc1 = rate / thick1. We also expect it to be negative and it tells how much percent thinning we have at a given cortical location.
The symmetrized percent change (spc) is the rate with respect to the average thickness: spc = rate / avg. This is a more robust measure than pc1, because thickness at time point 1 is more noisy than the average. Also spc is symmetric: when reversing the order of tp1 and tp2 it switches sign. This is not true for pc1. Therefore and for other reasons related to increased statistical power, we recommend to use spc.
So let's investigate (some of) the results, which can be found in each base surf directory. Call this to open up the pial surface (left hemi) of subject OAS2_0001:
tksurfer OAS2_0001 lh pial -overlay $SUBJECTS_DIR/OAS2_0001/surf/lh.long.thickness-avg.fwhm15.mgh -timecourse $SUBJECTS_DIR/OAS2_0001/surf/lh.long.thickness-stack.mgh -aparc
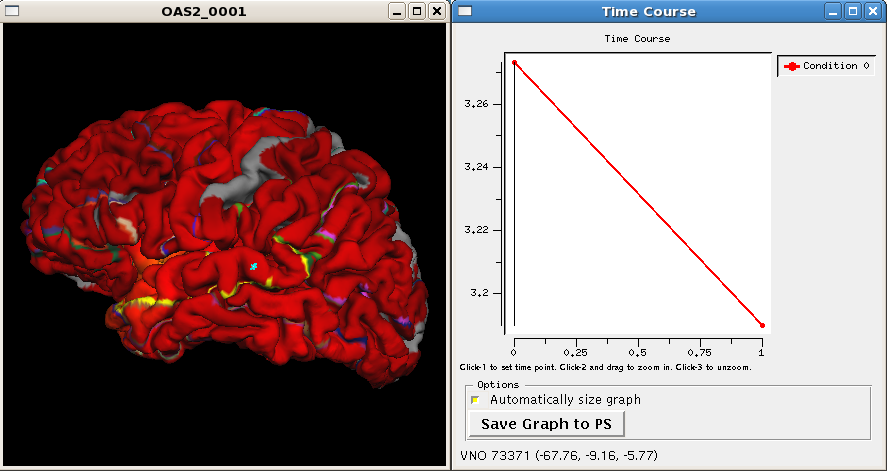
You are looking at the smoothed average thickness (color overlay). If you click at any location on the surface you can see a plot of the two original (unsmoothed) thickness values at the two time points. The -aparc flag opens the cortical parcellation which can be switched on and off, it helps to find out what region you are inspecting when clicking on the surface. For a single subject values are often noisy even after smoothing. That is why for a group analysis one needs several subjects in each group.
In a similar fashion you can open for example the symmetrized percent change. This time we open it on fsaverage, an average subject provided with FreeSurfer and often used as the common target to compare results across subjects. The --qcache flag of long_mris_slopes has conveniently registered and mapped all results to this average subject:
tksurfer fsaverage lh pial -overlay $SUBJECTS_DIR/OAS2_0001/surf/lh.long.thickness-spc.fwhm15.fsaverage.mgh -aparc
or if you prefer freeview (ignore the warning and click the 3D view icon):
freeview -f fsaverage/surf/lh.pial:overlay=$SUBJECTS_DIR/OAS2_0001/surf/lh.long.thickness-spc.fwhm15.fsaverage.mgh:overlay_threshold=2,5
note the 'fsaverage' in the filename. Because there is not much change on the surfaces, the spc will be even more noisy than the average.
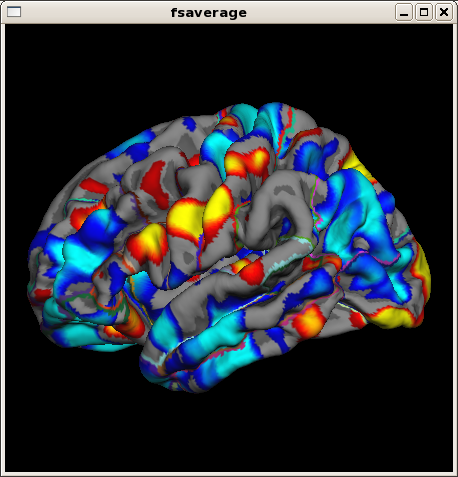
Since FS5.2: Similar to the thickness data, you can use long_stats_slopes to compute subject rates, pct. changes etc for the stats (aseg or aparc). Once you reduced the measures to a single number per subject, you can compare groups similar to a cross sectional analysis (e.g. using mri_glm_fit or qdec).
Additional Notes on QDEC
In order to run a QDEC group analysis (described in the QDEC tutorial), you need a qdec table. Since qdec cannot directly work with the longitudinal qdec tables yet, you have to shrink it into a cross sectional form. Use
long_qdec_table --qdec ./qdec/long.qdec.table.dat --cross --out ./qdec/cross.qdec.table.dat
to create a table with only a single line for each subject. For each subject, numerical values such as age or height will be averaged across time, other values will be copied from the first tp for each subject as ordered in the input table.
Also QDEC will not know about our new files (e.g. lh.long.thickness-spc...). We can tell it to look for them by creating a $SUBJECTS_DIR/qdec/.Qdecrc file in the qdec directory that contains the following lines:
MEASURE1 = long.thickness-avg MEASURE2 = long.thickness-rate MEASURE3 = long.thickness-pc1 MEASURE4 = long.thickness-spc
(If you are running your own data, make sure you create this file in the $SUBJECTS_DIR/qdec directory!)
You can then run qdec and do all kinds of analysis on any of those files and other variables from the qdec table:
qdec --table ./qdec/cross.qdec.table.dat
You will see in the Design tab, that you can now select e.g. long.thickness-rate under Measure as the dependent variable.
Editing Longitudinal Data
Editing longitudinal data can be complicated, but in some cases you actually save time, as some edits are only necessary in the base. You should be familiar with Edits and might want to check also the page about LongitudinalEdits.
Here are some examples of some common errors you can encounter with your data. Note: Because some of the recons take a while to run and it is not practical for you to run them during this short period of time, we ran the edits ahead of time. You will see that the edited and re-processed version have the postfix '_fixed'.
Before starting each section, remember to do the following to any new terminal you open:
setenv SUBJECTS_DIR $TUTORIAL_DATA/long-tutorial cd $SUBJECTS_DIR
tkmedit Cheat Sheet
(You can skip this now and look at if later when editing, if you forgot how to use these tools)
- Remember to set the Brush Info to either Main or Aux volume depending on which volume you want to edit. Ctrl + Left click on Edit Voxels tool will bring up the Brush Info and Volume Brush Info toolboxes. Do not make changes to the Volume Brush Info toolbox.
|
Key Shortcut |
Button 1 |
Button 2 |
Button 3 |
Navigation Tool |
n |
Clicking and dragging pans the view across the current slice. No effect when the zoom level is 1. |
Clicking once in the top half of the window increases the slice by 1, clicking in the bottom decreases the slice. Dragging up increases the slice, dragging down decreases it. |
Clicking once in the top half of the window zooms in, clicking in the bottom zooms out. Dragging up zooms in continuously, dragging down zooms out. |
Select Voxel Tool |
s |
Clicking sets the cursor. |
Clicking adds a voxel to the selection. |
Clicking removes a voxel from the selection. |
Edit Voxel Tool |
a |
Clicking sets the cursor. |
Clicking adds a voxel to the volume. |
Clicking removes a voxel from the volume. |
Edit Control Points Tool |
t |
Clicking sets the cursor. |
Clicking makes a new control point. |
Clicking removes nearest control point. |
Skullstrip Error
The most common type of skull strip failures you probably will see has to do with the cerebellum: Often, part of the cerebellum is stripped off (as you have seen in the skull strip tutorial in the troubleshooting portion earlier). Sometimes skull stripping will also leave too much dura behind. For either case, the best way to solve the problem is to adjust the watershed parameters.
Please take a look at subject OAS2_0088 in tkmedit.
tkmedit OAS2_0088 brainmask.mgz -aux T1.mgz -surfs
As always, you should also check out the cross-sectionals and longitudinals for the same subject to see if the problem (if there is one) is present in all parts of the streams.
To open the cross (in separate terminals, don't forget to set the SUBJECTS_DIR in a new terminal window):
tkmedit OAS2_0088_MR1 brainmask.mgz -aux T1.mgz -surfs tkmedit OAS2_0088_MR2 brainmask.mgz -aux T1.mgz -surfs
To open the longs (in separate terminals):
tkmedit OAS2_0088_MR1.long.OAS2_0088 brainmask.mgz -aux T1.mgz -surfs tkmedit OAS2_0088_MR2.long.OAS2_0088 brainmask.mgz -aux T1.mgz -surfs
These commands will open the brainmask.mgz volume, the T1.mgz loaded as aux, and the surfaces for both hemispheres. You can check the brainmask.mgz volume and compare it to the T1.mgz volume (use Alt+c to toggle back and forth between them).
As mentioned, skull strip failures are best fixed by adjusting the watershed parameters of the cross-sectionals (it can only be done at this part of the stream). Afterward, you can recreate the base and longitudinals. Did you find the problem? If so, feel free to run the -skullstrip step with a different watershed threshold for this subject. If you don't remember how to do it, click here for the detailed instructions, and also to see the edited version.
Pial/Brainmask Edits
Sometimes you will come across a case where you need to make edits to the brainmask.mgz to correct the pial surfaces. Open subject OAS2_0057 to see where and how this type of edits should be done.
First open the base.
tkmedit OAS2_0057 brainmask.mgz -aux T1.mgz -surfs
This will open the brainmask.mgz volume, T1.mgz volume as aux, and all surfaces. You should also check out the cross-sectionals and the longitudinals of this subject to check if the problem is present in all parts of the stream:
tkmedit OAS2_0057_MR1 brainmask.mgz -aux T1.mgz -surfs tkmedit OAS2_0057_MR2 brainmask.mgz -aux T1.mgz -surfs
tkmedit OAS2_0057_MR1.long.OAS2_0057 brainmask.mgz T1.mgz -surfs tkmedit OAS2_0057_MR2.long.OAS2_0057 brainmask.mgz T1.mgz -surfs
See if you can find the slices where you will need to edit the braimask.mgz in order to correct the surfaces. You can click here for a detailed description on how and at which stage you can fix this problem.
Control Points Edits
Sometimes, there are areas in the white matter with intensity lower than 110 and where the wm is not included in the surfaces. The best way to fix this problem is to add control points.
Take a look at the next subject, OAS2_0121 in all cross, base, and long to see if you can find the places that need control points.
tkmedit OAS2_0121_MR1 brainmask.mgz -aux T1.mgz -surfs tkmedit OAS2_0121_MR2 brainmask.mgz -aux T1.mgz -surfs
tkmedit OAS2_0121 brainmask.mgz -aux T1.mgz -surfs
tkmedit OAS2_0121_MR1.long.OAS2_0121 brainmask.mgz -aux T1.mgz -surfs tkmedit OAS2_0121_MR2.long.OAS2_0121 brainmask.mgz -aux T1.mgz -surfs
Click here if you want to see where to place the control points and what effects it has on the longitudinals.
White Matter Edits
White matter edits are possibly the most common type of edits you will come across in data. In many cases, even a difference of 1 wm voxel can cause a huge defect in the surfaces. Take a look at the following subject in tkmedit. For this one, we will open the brainmask.mgz volume and the wm.mgz volume as aux instead of T1.mgz.
tkmedit OAS2_0185 brainmask.mgz -aux wm.mgz -surfs
tkmedit OAS2_0185_MR1 brainmask.mgz -aux wm.mgz -surfs tkmedit OAS2_0185_MR2 brainmask.mgz -aux wm.mgz -surfs
tkmedit OAS2_0185_MR1.long.OAS2_0185 brainmask.mgz -aux wm.mgz -surfs tkmedit OAS2_0185_MR2.long.OAS2_0185 brainmask.mgz -aux wm.mgz -surfs
Toggle between the two volumes (use 'Alt-c') and see if you can spot an area where a simple wm edit will make a huge difference to the surfaces. Once you find it, or if you give up, click here to see how the edits can be done.
Brain.finalsurfs.manedit.mgz Edits
The brain.finalsurfs.manedit.mgz volume will allow for the correction of a particular type of problem involving pial surface misplacement whereby parts of the pial surface have extended into the cerebellum in certain areas. It is only in this case where the pial surface extends into cerebellum where the brain.finalsurfs.manedit.mgz volume should be edited. In all other non-cerebellum pial surface problems, brainmask.mgz should be edited. This cerebellum/pial surface problem can be fixed by removing those cerebellum voxels and other surrounding problematic voxels in the brain.finalsurfs.manedit.mgz volume such that upon running recon-all, the pial surface will be pulled in as desired, bordering only gray matter/ CSF boundary, and not jutting into the cerebellum. If only edits to this volume are made on a subject, it is sufficient to re-run recon-all from the point of these edits using the flag -autorecon3-pial. For the following subject (OAS2_0002), it is seen that both cross sectional time points (OAS2_0002_MR and MR2) have some errors of this nature in several slices of the brain.finalsurfs.mgz volume. In the corresponding region in the base (OAS2_0002), similar problems show up, and therefore, the longitudinal data sets for time points 1 and 2 also have some errors in the corresponding areas of the brain.finalsurfs.mgz volume. The tutorial below demonstrates how to manually fix the problem in the cross sectional time points and in the base, in order to automatically fix the problem in the long. Notice that edits will be made to a copy of the brain.finalsurfs.mgz file which will be named brain.finalsurfs.manedit.mgz. The tutorial walks you through editing cross-sectional time point 2 step by step, because the error in question is larger and better defined in this time point as compared to time point 1. However, the tutorial explains how to fix the problem in time point 1 aswell. In practice, you should check both cross-sectional time points for errors of this type, fix them and regenerate the crosssectionals. Then, you should check the base, fix any errors, and regenerate the base. Then, regenerate both longitudinal runs using the fixed cross-sectional data sets and the fixed base.
1. Look at the brain.finalsurfs.mgz volumes within OAS2_0002_MR2 at the slices containing cerebellum in order to see the problem.
tkmedit OAS2_0002_MR2 brainmask.mgz -aux brain.finalsurfs.mgz -surfs -segmentation aseg.mgz $FREESURFER_HOME/FreeSurferColorLUT.txt
Click here to view the problem and how to fix it.
2. Editing the Base: Open the base for this subject
tkmedit OAS2_0002 brainmask.mgz -aux brain.finalsurfs.mgz -surfs -segmentation aseg.mgz $FREESURFER_HOME/FreeSurferColorLUT.txt
Click here to view the problem and how to fix it.
3. Once the cross-sectional time points 1 and 2 and the base are fixed, it is time to recreate a long files for time points 1 and 2 (by renaming/removing the old and re-processing the -long from scratch). First you can view the original long file for time point 2 (for example) below:
tkmedit OAS2_0002_MR2.long.OAS2_0002 brainmask.mgz -aux brain.finalsurfs.mgz -surfs -segmentation aseg.mgz $FREESURFER_HOME/FreeSurferColorLUT.txt
Click here to see the results of creating a new long file from the edited base and cross-sectional. You will do the same for MR1.