|
Size: 2884
Comment: missing edit-log entry for this revision
|
Size: 5928
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [wiki:Self:FsTutorial top] | [wiki:Self:FsTutorial/FslFeatFreeSurfer previous] | [[FsTutorial|top]] | [[FsTutorial/FslFeatFreeSurfer|previous]] = Overlaying FSL Feat statistical maps = |
| Line 3: | Line 4: |
| == Overlaying FSL Feat statistical maps == | == Preparations == In this exercise, the data set of subject '''bert''' is used for demonstration. Type the following prior to beginning the exercises: === If You're at an Organized Course === If you are taking one of the formally organized courses, everything has been set up for you on the provided laptop. The only thing you will need to do is run the following commands in ''every'' '''new''' terminal window (aka shell) you open throughout this tutorial. Copy and paste the commands below to get started: |
| Line 5: | Line 9: |
| In this exercise, the data set of subject '''bert''' is used for demonstration. To begin the exercises, first enter the following directory, and then set the current directory to be the default subjects directory using this command: | |
| Line 7: | Line 10: |
| cd $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs setenv SUBJECTS_DIR ${PWD} |
setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR/fbert-feat ln -s $FREESURFER_HOME/subjects/fsaverage ln -s $FREESURFER_HOME/subjects/bert |
| Line 10: | Line 15: |
| ''To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.'' These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into this directory. You can now skip ahead to the tutorial (below the gray line). === If You're not at an Organized Course === If you are '''NOT''' taking one of the formally organized courses, then to follow this exercise exactly be sure you've downloaded the [[FsTutorial/Data|tutorial data set]] before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. These are the commands that you need to run before getting started: {{{ tcsh source your_freesurfer_dir/SetUpFreeSurfer.csh setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR/fbert-feat ln -s $FREESURFER_HOME/subjects/fsaverage ln -s $FREESURFER_HOME/subjects/bert }}} Notice the command to open tcsh. If you are already running the tcsh command shell, then the 'tcsh' command is not necessary. If you are not using the tutorial data you should set your {{{SUBJECTS_DIR}}} to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located. The subjects 'fsaverage' and 'bert' may already exist in the subjects directory, so ignore any warnings that might be issued that say 'File exists'. -------- |
|
| Line 13: | Line 37: |
| Use the following command to display the zmap (zstat1.img) overlaid onto the bert's orig volume. It will display the automatic segmentation, and will also set the threshold at z = 1.3: | Use the following command to display the zmap (zstat1.nii.gz) from the first run overlaid onto the bert's orig volume. It will display the automatic segmentation, and will also set the threshold at z = 2.3: ---- |
| Line 15: | Line 40: |
| tkmedit bert orig -aux brain -overlay ./fbert.feat/stats/zstat1.img \ -overlay-reg ./fbert.feat/reg/freesurfer/anat2exf.register.dat \ -fthresh 1.3 -fmid 2.3 -fslope 1 \ -segmentation ${SUBJECTS_DIR}/bert/mri/aseg ${FREESURFER_HOME}/tkmeditColorsCMA |
tkmedit bert orig.mgz lh.white -aux brain.mgz \ -overlay ./fbert1.feat/stats/zstat1.nii.gz \ -overlay-reg ./fbert1.feat/reg/freesurfer/anat2exf.register.dat \ -segmentation aparc+aseg.mgz -fthresh 2.3 -fmax 4.3 |
| Line 20: | Line 45: |
| ---- You should see the image below:<<BR>> {{attachment:tkm-zstat1-cor-128.th23.small.jpg}} <<BR>> When you click or mouse over a voxel, the cortical or subcortical structure that that voxel belongs to will be displayed in the control panel. You can view any of the volumes in the stats dir in this way as well as the clustered maps in the feat directory. |
|
| Line 21: | Line 50: |
| You should see the image below:[[BR]] attachment:tkm-zstat1-cor-128-small.jpg |
=== 2.0 View statistical maps on bert's surface === To view any of the statistical maps on bert's surface, close the tkmedit GUI (or open a new terminal window) and run: ---- {{{ tksurfer bert lh inflated \ -overlay ./fbert1.feat/stats/zstat1.nii.gz \ -overlay-reg ./fbert1.feat/reg/freesurfer/anat2exf.register.dat \ -fthresh 2.3 -fmid 3.3 -fslope 1 -annot aparc.annot }}} ---- Change the cortical parcellation to outline mode with View->LabelStyle->Outline. You should see the image below:<<BR>> {{attachment:tks-zstat1-rh-lat.th23.small.jpg}} <<BR>> When you click or mouse over a vertex, the control panel will display the name of the cortical structure. You can view any of the volumes in the stats dir in this way as well as the clustered maps in the feat directory. You can also run the tkmedit and tksurfer commands above in separate shells and use the Save-Point/Goto-Point functionality to navigate through the volume and surface. |
| Line 24: | Line 65: |
| === 2.0 Sampling statistical maps onto bert's surface === | === 3.0 Displaying Same-Subject, Cross-Run GFEAT Results === |
| Line 26: | Line 67: |
| In order to display the statistical map overlaid onto the surface, the data in fbert.feat must be resampled. This is done with feat2surf. Documentation for what it does and how it does it can be obtained with: | Typically, one collects more than one run/series of functional data for each subject. The individual runs are analyzed separately, then combined in standard space with GFEAT using a fixed-effects model. Since the data are no longer in the subject's native functional space, a different registration matrix is needed to map the GFEAT results to the individual. Each run of reg-feat2anat will create a reg/freesurfer/anat2std.register.dat. Any one of these can be used to map the GFEAT data to the subject's anatomy. First, verify that the registration is good with: ---- |
| Line 28: | Line 79: |
| feat2surf --help | tkregister2 --mov fbert.gfeat/mean_func.nii.gz --surf \ --reg fbert1.feat/reg/freesurfer/anat2std.register.dat |
| Line 30: | Line 82: |
| To run it on this data set, run: | ---- mean_func.nii.gz is the mean of the example_func's in standard space. Since this is the mean func, it will be blurry if you have spatially smoothed. Note: if there is a problem with this registration, you need to repair the registrations for each individual run. Now show gfeat results on anatomical volume: ---- |
| Line 32: | Line 92: |
| feat2surf --feat fbert.feat | tkmedit bert orig.mgz -seg aparc+aseg.mgz \ -ov fbert.gfeat/cope1.feat/stats/zstat1.nii.gz \ -ovreg fbert1.feat/reg/freesurfer/anat2std.register.dat \ -fthresh 2.3 -fmax 4.3 |
| Line 34: | Line 97: |
| Verify that this has created four directories under fbert.feat: reg_surf-lh, reg_surf-rh, reg_surf-lh-average7, and reg_surf-rh-average7. Each one of these directories will have a stats directory in which all the statistics from fbert.feat/stats have been resampled onto the surface. The average7 directories are resampled onto the common surface spaced (defined by average7). | ---- Here we've used the anat2std.register.dat from the first run. |
| Line 36: | Line 100: |
| === 3.0 Viewing statistical maps on bert's surface === To view the statistical maps on the right hemisphere, run tksurfer: |
Now show gfeat results on the surface: ---- |
| Line 40: | Line 103: |
| tksurfer bert rh inflated \ -o ./fbert.feat/reg_surf-rh/stats/zstat1.mgh -overlay-reg-identity \ -fthresh 1.3 -fmid 2.3 -fslope 1 |
tksurfer bert lh inflated -annot aparc.annot \ -ov fbert.gfeat/cope1.feat/stats/zstat1.nii.gz \ -ovreg fbert1.feat/reg/freesurfer/anat2std.register.dat \ -fthresh 2.3 -fmid 3.3 -fslope 1 |
| Line 44: | Line 108: |
| Note that you can run the above tkmedit command (from a different window) simultaneously with the tksurfer command to visualize the same data on the volume and in the surface. You can then use the Save/Goto Point buttons to navigate between the surface and volume. You should see the image below:[[BR]] attachment:tks-zstat1-rh-lat-small.jpg === 4.0 Overlaying zmap onto the FSL's standard volume === You can also use tkmedit to view your results on the FSL standard volume instead of the individual's anatomical with the following command: {{{ tkmedit -f $FSLDIR/etc/standard/avg152T1_brain.img \ -overlay ./fbert.feat/stats/zstat1.img \ -overlay-reg ./fbert.feat/reg/freesurfer/std2exf.register.dat \ -fthresh 1.3 -fmid 2.3 -fslope 1 }}} |
---- |
Overlaying FSL Feat statistical maps
Preparations
In this exercise, the data set of subject bert is used for demonstration. Type the following prior to beginning the exercises:
If You're at an Organized Course
If you are taking one of the formally organized courses, everything has been set up for you on the provided laptop. The only thing you will need to do is run the following commands in every new terminal window (aka shell) you open throughout this tutorial. Copy and paste the commands below to get started:
setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR/fbert-feat ln -s $FREESURFER_HOME/subjects/fsaverage ln -s $FREESURFER_HOME/subjects/bert
To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.
These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into this directory. You can now skip ahead to the tutorial (below the gray line).
If You're not at an Organized Course
If you are NOT taking one of the formally organized courses, then to follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. These are the commands that you need to run before getting started:
tcsh source your_freesurfer_dir/SetUpFreeSurfer.csh setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR/fbert-feat ln -s $FREESURFER_HOME/subjects/fsaverage ln -s $FREESURFER_HOME/subjects/bert
Notice the command to open tcsh. If you are already running the tcsh command shell, then the 'tcsh' command is not necessary. If you are not using the tutorial data you should set your SUBJECTS_DIR to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located.
The subjects 'fsaverage' and 'bert' may already exist in the subjects directory, so ignore any warnings that might be issued that say 'File exists'.
1.0 Overlaying the statistical map onto the bert's orig volume
Use the following command to display the zmap (zstat1.nii.gz) from the first run overlaid onto the bert's orig volume. It will display the automatic segmentation, and will also set the threshold at z = 2.3:
tkmedit bert orig.mgz lh.white -aux brain.mgz \ -overlay ./fbert1.feat/stats/zstat1.nii.gz \ -overlay-reg ./fbert1.feat/reg/freesurfer/anat2exf.register.dat \ -segmentation aparc+aseg.mgz -fthresh 2.3 -fmax 4.3
You should see the image below:
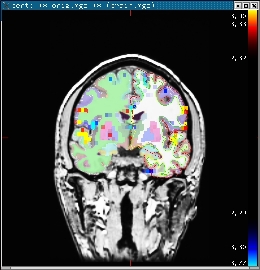
When you click or mouse over a voxel, the cortical or subcortical structure that that voxel belongs to will be displayed in the control panel. You can view any of the volumes in the stats dir in this way as well as the clustered maps in the feat directory.
2.0 View statistical maps on bert's surface
To view any of the statistical maps on bert's surface, close the tkmedit GUI (or open a new terminal window) and run:
tksurfer bert lh inflated \ -overlay ./fbert1.feat/stats/zstat1.nii.gz \ -overlay-reg ./fbert1.feat/reg/freesurfer/anat2exf.register.dat \ -fthresh 2.3 -fmid 3.3 -fslope 1 -annot aparc.annot
Change the cortical parcellation to outline mode with View->LabelStyle->Outline. You should see the image below:
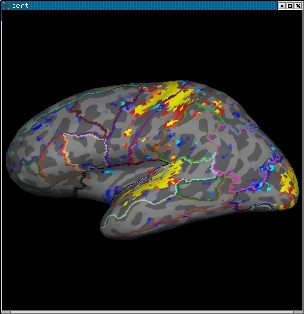
When you click or mouse over a vertex, the control panel will display the name of the cortical structure. You can view any of the volumes in the stats dir in this way as well as the clustered maps in the feat directory. You can also run the tkmedit and tksurfer commands above in separate shells and use the Save-Point/Goto-Point functionality to navigate through the volume and surface.
3.0 Displaying Same-Subject, Cross-Run GFEAT Results
Typically, one collects more than one run/series of functional data for each subject. The individual runs are analyzed separately, then combined in standard space with GFEAT using a fixed-effects model. Since the data are no longer in the subject's native functional space, a different registration matrix is needed to map the GFEAT results to the individual. Each run of reg-feat2anat will create a reg/freesurfer/anat2std.register.dat. Any one of these can be used to map the GFEAT data to the subject's anatomy.
First, verify that the registration is good with:
tkregister2 --mov fbert.gfeat/mean_func.nii.gz --surf \ --reg fbert1.feat/reg/freesurfer/anat2std.register.dat
mean_func.nii.gz is the mean of the example_func's in standard space. Since this is the mean func, it will be blurry if you have spatially smoothed. Note: if there is a problem with this registration, you need to repair the registrations for each individual run.
Now show gfeat results on anatomical volume:
tkmedit bert orig.mgz -seg aparc+aseg.mgz \ -ov fbert.gfeat/cope1.feat/stats/zstat1.nii.gz \ -ovreg fbert1.feat/reg/freesurfer/anat2std.register.dat \ -fthresh 2.3 -fmax 4.3
Here we've used the anat2std.register.dat from the first run.
Now show gfeat results on the surface:
tksurfer bert lh inflated -annot aparc.annot \ -ov fbert.gfeat/cope1.feat/stats/zstat1.nii.gz \ -ovreg fbert1.feat/reg/freesurfer/anat2std.register.dat \ -fthresh 2.3 -fmid 3.3 -fslope 1
