Segmentation of hypothalamic subunits
This functionality is available in FreeSurfer 7.2.
Author: Benjamin Billot
E-mail: benjamin.billot [at] inria.fr
Rather than directly contacting the author, please post your questions on this module to the FreeSurfer mailing list at freesurfer [at] nmr.mgh.harvard.edu
If you use these tools in your analysis, please cite:
Automated segmentation of the hypothalamus and associated subunits in brain MRI. Billot, B., Bocchetta, M., Todd, E., Dalca, A.V., Rohrer, J.D., Iglesias, J.E. Neuroimage (in press).
Contents
- General Description
- Installation
- Usage
- Frequently asked questions
1. General Description
This tool enables automated segmentation of the hypothalamus and its associated subunits in 3D T1-weighted scans of approximately 1mm isotropic resolution. It uses a convolutional neural network, which enables computing segmentations in a very short processing time (around 10 seconds with a CPU, less than a second with a GPU). More specifically, this code produces segmentation maps for five subregions (with distinct right/left labels), which comprise the following hypothalamic nuclei:
Subunits |
Left label |
Right label |
Associated nuclei |
Anterior-inferior |
801 |
806 |
suprachiasmatic nucleus; supraoptic nucleus (SON) |
Anterior-superior |
802 |
807 |
preoptic area; paraventricular nucleus (PVN) |
Posterior |
803 |
808 |
mamillary body (including medial and lateral mamillary nuclei); lateral hypothalamus; tuberomamillary nucleus (TMN) |
Inferior tubular |
804 |
809 |
infundibular (or arcuate) nucleus; ventromedial nucleus; SON; lateral tubular nucleus; TMN |
Superior tubular |
805 |
810 |
dorsomedial nucleus; PVN; lateral hypothalamus |
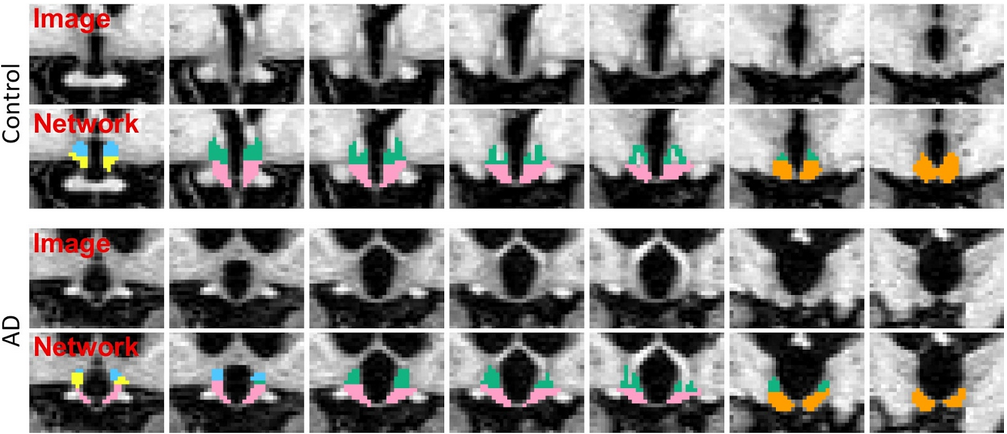
The segmentation model was trained by applying aggressive data augmentation, which makes it robust against variability in acquisition parameters (sequence, platform, bias field, head positioning), and in anatomy (e.g. atrophy patterns linked to ageing or different pathologies). The following figure illustrates two segmentation examples in coronal slices obtained by this model for a control subject, and a subject diagnosed with Alzheimer's disease:
2. Installation
The first time you run this module, it will prompt you to install Tensorflow. Simply follow the instructions in the screen to install the CPU or GPU version.
If you have a compatible GPU, you can install the GPU version for faster processing, but this requires installing libraries (GPU driver, Cuda, CuDNN). These libraries are generally required for a GPU, and are not specific for this tool. In fact you may have already installed them. In this case you can directly use this tool without taking any further actions, as the code will automatically run on your GPU.
3. Usage
This software can be used in two modes:
on subjects processed with FreeSurfer
- on any T1 scan of approximately 1mm resolution.
The first mode follows the FreeSurfer subject directory structure and works on the bias field corrected images (nu.mgz). We recommend using this mode if you are running the main FreeSurfer recon-all scripts on your data. The second mode is more flexible and has the advantage of not having to run subjects through recon-all, if you are only interested in the hypothalamus.
Important: the network has been trained on image at approximately 1mm isotropic resolution, so it would be underperforming if presented with images of higher/lower resolutions. Depending on the version of FreeSurfer (but regardless of the used mode, i.e., FreesSurfer or any-T1), there are several options:
in the development version any image with a resolution outside the [0.95, 1.15] range will be internally resampled to 1mm resolution, regardless of the selected mode (i.e. FreeSurfer subjects or any-T1).
for all the FreeSurfer versions equal or lower than 7.2, this resamping will have to be done externally, for example with the FreeSurfer command mri_convert:
mri_convert <path input image> <path resampled image> --voxsize 1 1 1 -odt float
We now describe what command to use in the different modes. A summary of the following information can be found simply by typing:
mri_segment_hypothalamic_subunits -h
a. FreeSurfer subjects
This tool can be run on subjects that have been analyzed beforehand with the main FreeSurfer stream (i.e. “recon-all”). Therefore, we will assume that a command similar to the following one has already been run on each subject for which we want to segment the hypothalamic subunits:
recon-all -all --s <subject_name>
To analyze one or several subjects, simply use:
mri_segment_hypothalamic_subunits --s <subject_name> --sd <subjects_dir> --write_posteriors --threads <threads> --cpu
where:
<subject_name>: is the name of a subject in $SUBJECTS_DIR. You can also specify several subjects to run them all at the same time. If no name is given after --s, the code will be run on ALL subjects in $SUBJECTS_DIR.
<subjects_dir>: (optional) is the path to a folder with which to override the current value of $SUBJECTS_DIR.
--write_posteriors: (optional) will save the posteriors (soft segmentations) obtained for each subject. This is not recommended unless needed, since it greatly increases the processing time.
<threads>: (optional) number of threads to be used by Tensorflow (default uses one core). Increase it to decrease the runtime when using the CPU version.
--cpu: (optional) use this flag to enforce running with CPU rather than GPU.
When running on a subject analysed with FreeSurfer, this software automatically saves the segmentation and volumes of the hypothalamic subunits in the FreeSurfer directory structure.
Segmentations are saved under <subjects_directory>/<subject_name>/mri/hypothalamic_subunits_seg.v1.mgz.
(in the dev version of FreeSurfer) If the input image had to be resampled to 1mm resolution, then the resample image is also saved under <subjects_directory>/<subject_name>/mri/hypothalamic_subunits_nu_resampled_1mm.v1.mgz.
Volumes are saved under <subjects_directory>/<subject_name>/mri/hypothalamic_subunits_volumes.v1.csv.
Volumetric stats are saved under <subjects_directory>/<subject_name>/stats/hypothalamic_subunits_volumes.v1.stats.
Posteriors (when enabled) are saved under <subjects_directory>/<subject_name>/mri/hypothalamic_subunits_posteriors.v1.mgz.
Important: if several subjects are processed, the outputs will be saved in each subject's directory. Additionally, a CSV file regrouping the volumes of all structures for all subjects will be saved under:
- $SUBJECTS_DIR/hypothalamic_subunits_volumes_all.v1.csv.
Examples:
Segment all subjects in $SUBJECTS_DIR:
mri_segment_hypothalamic_subunits --s
Segment a single subject bert:
mri_segment_hypothalamic_subunits --s bert
Segment several subjects (bert and ernie):
mri_segment_hypothalamic_subunits --s bert ernie
Segment a single subject bert located in a subject directory other than $SUBJECTS_DIR:
mri_segment_hypothalamic_subunits --s bert --sd /home/user1/data/new_analysis/
Segment all subjects located in a subject directory other than $SUBJECTS_DIR:
mri_segment_hypothalamic_subunits --s --sd /home/user1/data/new_analysis/
b. Any T1 scan
You can also run this on a T1 scan that has not been processed with FreeSurfer. This tool works on scans with or without preprocessing (e.g., bias field correction, skull stripping, normalization, template registration, etc.). Depending on the version of FreeSurfer you use, there are two ways to call mri_segment_hypothalamic_subunits.
For FreeSurfer 7.2 or lower, the command line is:
mri_segment_hypothalamic_subunits --i <input> --o <output> [--out_posteriors <post> --out_volumes <vol> --threads <threads>]
where:
<input>: is the path to a scan to segment. This can also be a folder, in which case all the scans in this folder will be segmented.
<output>: is the path where the output segmentation will be saved. This must be a folder if <input> is a folder.
<post>: (optional) is the path where the posteriors (given as soft probability maps) will be saved. This must also be a folder if <input> is a folder.
<vol>: (optional) is the path to an output CSV file where the volumes of the whole hypothalamus and of all subunits will be saved. In the case where <input> designates a folder, the volumes of all subjects will be saved in the same file.
<threads>: (optional) number of threads to be used by Tensorflow (default uses one core). Increase it to decrease the runtime when using the CPU version.
Important: If you wish to process several scans in the any-T1 mode, we highly recommend that you put them in a single folder, rather than calling mri_segment_hypothalamic_subunits individually on each scan. This will save the time required to set up the software for each scan.
For the dev version, the command line is:
mri_segment_hypothalamic_subunits --i <input> --o <output> [--post <post> --resample <resample> --vol <vol> --threads <threads> --cpu --crop <crop>]
where:
<input>: is the path to a scan to segment. This can also be a folder, in which case all the scans in this folder will be segmented.
<output>: is the path where the output segmentation will be saved. This must be a folder if <input> is a folder.
<post>: (optional) is the path where the posteriors (given as soft probability maps) will be saved. This must also be a folder if <input> is a folder.
<resample>: (optional) in order to return segmentations at 1mm resolution, the input images are internally resampled to this resolution (except if they have a resolution in the range [0.95, 1.15]). Use this optional flag to save the resampled images. This must be a folder if <input> is a folder.
<vol>: (optional) is the path to an output CSV file where the volumes of the whole hypothalamus and of all subunits will be saved. In the case where <input> designates a folder, the volumes of all subjects will be saved in the same file.
<threads>: (optional) number of threads to be used by Tensorflow (default uses one core). Increase it to decrease the runtime when using the CPU version.
--cpu: (optional) use this flag to enforce running with CPU rather than GPU.
<crop>: (optional) to crop the inputs to a given shape before segmentation. This must be divisible by 8. It can be given as a single (i.e., --crop 160) or several integers (i.e, --crop 160 128 184, ordered in RAS coordinates). This value defaults to 184, decreased it for faster analysis or to fit in your GPU.
Important 1: If you wish to process several scans in the any-T1 mode, we highly recommend that you put them in a single folder, rather than calling mri_segment_hypothalamic_subunits individually on each scan. This will save the time required to set up the software for each scan.
Important 2: Because hypothalamus_seg may resample the images at 1mm isotropic resolution, some viewers will not display them correctly when overlaying the segmentations on the original images. If that’s the case, you can use the --resample flag to obtain a resampled image that lives in the same space as the segmentation, such that any viewer can be used to visualize them together.
Examples:
Segment a single scan:
mri_segment_hypothalamic_subunits --i /home/user1/data/scan.mgz --o /home/user1/data/seg.mgz
Segment a single scan, and save the resampled image and volumes:
mri_segment_hypothalamic_subunits --i /home/user1/data/scan.mgz --o /home/user1/data/seg.mgz --resample /home/user1/data/image_1mm3.mgz --vol /home/user1/data/hypothalamic_volumes.csv
Segment all scans contained in a folder, and save the posteriors and the volumes:
mri_segment_hypothalamic_subunits --i /home/user1/data/scans/ --o /home/user1/data/segmentations/ --post /home/user1/data/posteriors/ --vol /home/user1/data/hypothalamic_volumes.csv
4. Frequently asked questions (FAQ)
The results are bad, what can I do about it?
SO FAR, EXPERIENCE SHOWS THAT ALL BAD RESULTS ARE DUE TO INPUT SCANS AT RESOLUTION DIFFERENT THAN 1MM. That is actually why we implemented the internal resampling in the dev version. So if you're using FreeSurfer 7.2 or lower, please make sure that the input scans are at 1mm isotropic resolution. If the problem still persists, or if the results are bad and you use the dev version, please let us know, we'd be delighted to help you and to make our tool stronger ! ![]()
Does running this tool require preprocessing of the input scans?
No! Because we applied aggressive augmentation during training (see paper), this tool is able to segment both processed and unprocessed data. So there is no need to apply bias field correction, skull stripping, or intensity normalization.
The sum of the number of voxels of a given structure multiplied by the volume of a voxel is not equal to the volume reported in the output volume file.
This is because the volumes are computed upon a soft segmentation, rather than the discrete segmentation. The same happens with the main recon-all stream: if you compute volumes by counting voxels in aseg.mgz, you don't get the values reported in aseg.stats.
What formats are supported in the second mode (any-T1)?
This tool can be run on Nifti (.nii/.nii.gz) and FreeSurfer (.mgz) scans.
Which scan is actually used when inputting a FreeSurfer subject?
In this case, the module segments nu.mgz, which is corrected for bias field.
How can I increase the speed of the CPU version without using a GPU?
If you have a multi-core machine, you can increase the number of threads with the --threads flag (up to the number of cores). Additionally you can also try to decrease the cropping value, but this will also decrease the field of view of the image.
Why are the input images sometimes resampled to 1mm resolution? (dev version)
This is because the network has been trained to segment at this resolution, and could be underperforming if presented with images of different resolutions. Actually, only the images with resolutions outside the range [0.95, 1.15] are resampled. We highlight that the resampling is performed internally to avoid the dependence on any external tool.
Why aren't the segmentations perfectly aligned with their corresponding images? (dev version)
This is probably of problem of image viewer, in the case where the images have been resampled to 1mm resolution. Indeed, if images are resampled, their segmentations are given at 1mm resolution (so not the same as the input image), and some viewers cannot cope with resolution changes. In that case we recommend to use the --resample flag to save the resamplde images, or simply to use FreeView (shipped with FreeSurfer), which is resolution-aware !
