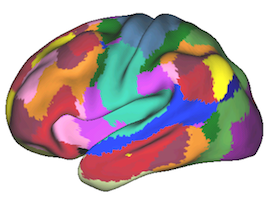
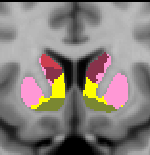
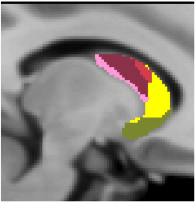
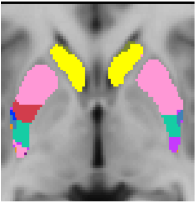
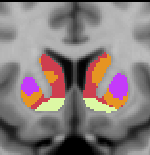
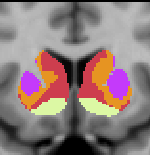
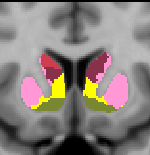
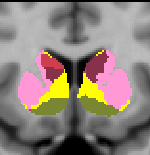
Striatal Parcellation Estimated by Intrinsic Functional Connectivity
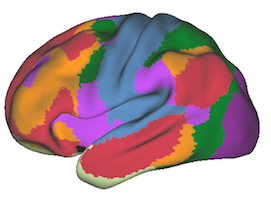
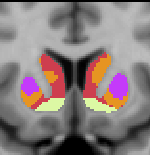
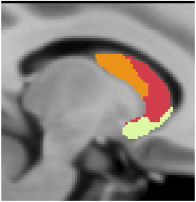
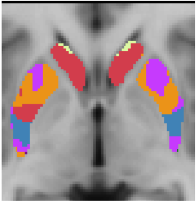
Data from 1000 young healthy adults were registered using nonlinear deformation of the striatum in combination with surface-based alignment of the cerebral cortex. All data were acquired on Siemens 3T scanners using the same functional and structural sequences. A clustering approach was employed to identify and replicate networks of functionally coupled regions across the cerebral cortex (see Yeo et al. 2011). The complete topography of the striatum was then mapped by estimating the principal cerebral target for each voxel in the striatum (see Choi et al. 2012 for more details). The parcellations agree with prior models based on monkey anatomy that divide the striatum into broad functional territories of reward, cognition, and motor function. The majority of the human striatum maps to cerebral association areas, with distinct zones of the striatum coupling to the distributed regions that comprise each large-scale associationetwork.
The striatal volumes in which each striatal voxel is assigned to a cerebral network (using either the 7 or 17 network solution in Yeo et al. 2011) are available for download here.
References
Striatal Parcellations in Nonlinear MNI152 Space
7 Network Estimate - Cerebral Cortex |
7 Network Estimate - Striatum |
|
|
|
|
|
|
17 Network Estimate - Cerebral Cortex |
17 Network Estimate - Striatum |
|
|
|
|
|
|
Downloads
Resting State Striatal Parcellation in FSL MNI152 space. (2.2 Mb download).
Information about Downloads
1. FSL_MNI152_FreeSurferConformed_1mm.nii.gz is the FSL MNI152 1mm template interpolated and intensity normalized into a 256 x 256 x 256 1mm-isotropic volume (obtained by putting the FSL MNI152 1mm template through recon-all using FreeSurfer 4.5.0)
2. Choi2012_7Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz is a volume where each striatal voxel was assigned to the cerebral network (out of 7 cerebral networks estimated in Yeo et al., 2011) with the most similar pattern of connectivity.
3. Choi2012_7Networks_MNI152_FreeSurferConformed1mm_TightMask.nii.gz is a volume where each striatal voxel was assigned to the cerebral network (out of 7 cerebral networks estimated in Yeo et al., 2011) with the most similar pattern of connectivity. Here, the striatal mask is tighter than in (2). The slices of this volume are shown in Choi et al., 2012.
4. Choi2012_7NetworksConfidence_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz displays the confidence of the network assignment in Choi2012_7Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz.
5. Choi2012_7NetworksConfidence_MNI152_FreeSurferConformed1mm_TightMask.nii.gz displays the confidence of the network assignment in Choi2012_7Networks_MNI152_FreeSurferConformed1mm_TightMask.nii.gz.
6. Choi2012_7Networks_ColorLUT.txt is a FreeSurfer readable text file specifying how the 7 networks are named, numbered, and colored in Choi et al., 2012:
Index
Network Name
R
G
B
0
NONE
0
0
0
0
1
7Networks_1
120
18
134
0
2
7Networks_2
70
130
180
0
3
7Networks_3
0
118
14
0
4
7Networks_4
196
58
250
0
5
7Networks_5
220
248
164
0
6
7Networks_6
230
148
34
0
7
7Networks_7
205
62
78
0
In particular the networks are numbered from 7Networks_1 to 7Networks_7. The first column of the text file specifies the value of voxels in the nifti values corresponding to the particular network. The second column of the text file specifies the name of the networks. For example, from the text file, voxels whose values = 3 corresponds to the network 7Networks_3. Columns 3 to 5 correspond to the R, G, B values (ranges from 0 to 55) of the networks. Last column is all zeros (FreeSurfer's default).
7. Choi2012_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz is a volume where each striatal voxel was assigned to the cerebral network (out of 17 cerebral networks estimated in Yeo et al., 2011) with the most similar pattern of connectivity.
8. Choi2012_17Networks_MNI152_FreeSurferConformed1mm_TightMask.nii.gz is a volume where each striatal voxel was assigned to the cerebral network (out of 17 cerebral networks estimated in Yeo et al., 2011) with the most similar pattern of connectivity. Here, the striatal mask is tighter than in (7). The slices of this volume are shown in Choi et al., 2012.
9. Choi2012_17NetworksConfidence_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz displays the confidence of the network assignment in Choi2012_17Networks_MNI152_FreeSurferConformed1mm_LooseMask.nii.gz.
10. Choi2012_17NetworksConfidence_MNI152_FreeSurferConformed1mm_TightMask.nii.gz displays the confidence of the network assignment in Choi2012_17Networks_MNI152_FreeSurferConformed1mm_TightMask.nii.gz.
11. Choi2012_17Networks_ColorLUT.txt is a FreeSurfer readable text file specifying how the 17 networks are named, numbered, and colored in Choi et al., 2012:
Index
Network Name
R
G
B
0
NONE
0
0
0
0
1
17Networks_1
120
18
134
0
2
17Networks_2
255
0
0
0
3
17Networks_3
70
130
180
0
4
17Networks_4
42
204
164
0
5
17Networks_5
74
155
60
0
6
17Networks_6
0
118
14
0
7
17Networks_7
196
58
250
0
8
17Networks_8
255
152
213
0
9
17Networks_9
200
248
164
0
10
17Networks_10
122
135
50
0
11
17Networks_11
119
140
176
0
12
17Networks_12
230
148
34
0
13
17Networks_13
135
50
74
0
14
17Networks_14
12
48
255
0
15
17Networks_15
0
0
130
0
16
17Networks_16
255
255
0
0
17
17Networks_17
205
62
78
0
Example Usage
See README in unzipped folder.
7 Network Tight Mask |
7 Network Liberal Mask |
17 Network Tight Mask |
17 Network Liberal Mask |
|
|
|
|
1) Except for the colortables, all the volumes are nifti volumes which can be read using any software like freeview (FreeSurfer), fslview (FSL), etc.
2) To overlay the 7-network volume on the FSL MNI152 1mm template in freeview (assuming FreeSurfer is already installed) with the appropriate colors, type the following assuming the working directory is in the same directory as the README:
freeview -v FSL_MNI152_FreeSurferConformed_1mm.nii.gz Choi2012_7Networks_MNI152_FreeSurferConformed1mm_TightMask.nii.gz:colormap=lut:lut=Choi2012_7Networks_ColorLUT.txt
3) To overlay the 7-network volume and the confidence of the network assignments on the FSL MNI152 1mm template in freeview (assuming FreeSurfer is already installed) with the appropriate colors, type the following assuming the working directory is in the same directory as the README:
freeview -v FSL_MNI152_FreeSurferConformed_1mm.nii.gz Choi2012_7Networks_MNI152_FreeSurferConformed1mm_TightMask.nii.gz:colormap=lut:lut=Choi2012_7Networks_ColorLUT.txt Choi2012_7NetworksConfidence_MNI152_FreeSurferConformed1mm_TightMask.nii.gz:colormap=heat:heatscale=0,0.5,1
Other downloads
Cerebral Surface Parcellations in FreeSurfer Space
Cerebral Surface Parcellations in FSL MNI152 1mm Volumetric Space
Cerebral Surface Parcellations in Caret Space
Movies of Striatal Seed-based Functional Connectivity