top | previous| next (Directory Structure)
Contents
1. FSFAST Tutorial Data Description
The functional data were collected as part of the Functional Biomedical Research Network (fBIRN, www.nbirn.net).
- Working-memory paradigm with distractors
- 18 subjects
- Each subject has 1 run (except sess01 which has 4 runs)
- Collected at MGH Bay 4 (3T Siemens)
FreeSurfer anatomical analyses
2. Functional Paradigm
The paradigm was designed to study the effects of emotional stimuli on the ability to recall items stored in working memory.
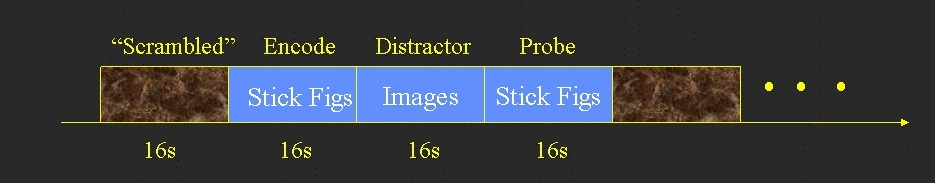
- Blocked design
- Each block consisted of 3 phases
- Encode (16 sec) - 8 stick figures to remember (no response)
- Distractor (16 sec) - 8 distractor images (response whether there is a face in the image)
- Emotional - Distractors are emotionally disturbing
- Neutral - Distractors are emotionally neutral
- Probe (16 sec) - 8 pairs of stick figures. Subject responds as to which of the pair was in the original Encode.
- Between each block was a 16 sec scrambled image used as baseline.
 The above yields 5 conditions:
The above yields 5 conditions:
- Encode
- Emotional Distractor
- Neutral Distractor
- Probe following Emotional Distractor
- Probe following Neutral Distractor
The scrambled image will be modeled as a baseline, not as a condition.
3. Functional Data
- Original data: each subject had 8 runs
- This data: each subject has 1 run (except for sess01 who has 4)
- Each run lasts 142 time points
- TR = 2 sec.
- There is one run of rest data for 13 subjects
- There is a B0 map for each subject
4. Anatomical Data
FreeSurfer analysis has been run for all 18 subjects
If you are at a FreeSurfer Course, continue on to the next page now
top | previous| next (Directory Structure)
5. Getting the Data (not necessary for the Boston FreeSurfer Course)
You can get the analyzed functional data (10G) from:
wget ftp://surfer.nmr.mgh.harvard.edu/pub/data/fsfast-functional.tar.gz
You can get the structural data (5G) from:
wget ftp://surfer.nmr.mgh.harvard.edu/pub/data/fsfast-tutorial.subjects.tar.gz
6. Organizing the Tutorial (not necessary for the Boston FreeSurfer Course)
cd to a place on your network where you have enough space to unpack the tutorial data.
cd /place/with/space
Untar the data
tar xvfz fsfast-tutorial.tar.gz tar xvfz fsfast-tutorial.subjects.tar.gz
You will need to set the TUTORIAL_DATA environment variable. In tcsh or csh
export TUTORIAL_DATA=/place/with/space
You will also need to link the FreeSurfer anatomical subjeccts (data in fsfast-tutorial.subjects) into your $SUBJECTS_DIR. You should set the FSFAST output format to be compressed NIFTI (nii.gz):
export FSF_OUTPUT_FORMAT=nii.gz
