|
Size: 1652
Comment:
|
← Revision 29 as of 2021-09-22 11:38:51 ⇥
Size: 3786
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [wiki:Self:FsTutorial top] | [wiki:Self:FsTutorial/GroupAnalysis previous] | [[FsTutorial|top]] | [[FsTutorial/GroupAnalysis|previous]] |
| Line 4: | Line 4: |
| *To follow this exercise exactly be sure you've downloaded the [[FsTutorial/Data|tutorial data set]] before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. | |
| Line 5: | Line 6: |
| In order to compute the contrats you will first need to change to the tutorial data directory and setup SUBJECTS_DIR: | In order to compute the contrasts you will first need to change to the tutorial data directory and setup SUBJECTS_DIR: |
| Line 8: | Line 9: |
| cd $FREESURFER_HOME/subjects/buckner_data/tutorial_subjs/group_analysis_tutorial | cd $TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial |
| Line 18: | Line 19: |
| Upon successful completion of the make_average_subject command you should have an average subject in your SUBJECTS_DIR. After successfully completing [wiki:Self:FsTutorial/CreateFsgdFile Exercise A.] and [wiki:Self:FsTutorial/CreateContrastVectors Exercise C.] you should have an FSGD file called my_gender_age.txt and a contrast file called age.mat, both in your SUBJECTS_DIR/stats directory . The following are sample commands, that can be used with the data, to complete a group analysis: | Upon successful completion of the make_average_subject command you should have an average subject in your SUBJECTS_DIR. After successfully completing [[FsTutorial/CreateFsgdFile|Exercise A.]] and [[FsTutorial/CreateContrastVectors|Exercise B.]] you should have an FSGD file called my_gender_age_fsgd.txt and a contrast file called age.mat, both in your SUBJECTS_DIR/stats directory. Confirm that those files exist: {{{ ls my_gender_age_fsgd.txt age.mat }}} If my_gender_age_fsgd.txt does not exist, complete [[FsTutorial/CreateFsgdFile|Exercise A.]] If age.mat does not exist, complete [[FsTutorial/CreateContrastVectors|Exercise B.]], or just create a file called age.mat containing this line: {{{ 0 0 1 }}} The following are sample commands, that can be used with the data, to complete a group analysis: |
| Line 23: | Line 33: |
| mris_preproc --fsgd my_gender_age.txt --target average --hemi lh -- meas thickness --out lh.my_gender_age.thickness.mgh | mris_preproc --fsgd my_gender_age_fsgd.txt \ --target average \ --hemi lh \ --meas thickness \ --out lh.my_gender_age.thickness.mgh |
| Line 26: | Line 40: |
Once this completes, you can do the smoothing step: |
Once this completes you should see the file lh.my_gender_age.thickness.mgh. The next step, the smoothing step, will use this as input: |
| Line 30: | Line 43: |
| mri_surf2surf --hemi lh --s average --sval lh.my_gender_age.thickness.mgh --fwhm 10 --tval lh.my_gender_age.thickness.10.mgh | mri_surf2surf --hemi lh \ --s average \ --sval lh.my_gender_age.thickness.mgh \ --fwhm 10 \ --tval lh.my_gender_age.thickness.10.mgh |
| Line 33: | Line 50: |
| Once this is complete, you can move on to test your contrast vector: | Once this is complete you should see the file lh.my_gender_age.thickness.10.mgh. The next step will test the hypothesis you've set up using your contrast vector: |
| Line 36: | Line 53: |
| mri_glmfit --y lh.my_gender_age.thickness.10.mgh --fsgd my_gender_age.txt --glmdir lh.my_gender_age.glmdir --pca --surf --average lh --C age.mat | mri_glmfit --y lh.my_gender_age.thickness.10.mgh \ --fsgd my_gender_age_fsgd.txt doss \ --glmdir lh.my_gender_age.glmdir \ --pca \ --surf average lh \ --C age.mat |
| Line 39: | Line 61: |
| You should have two new directories in $TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial/stats - lh.my_gender_age.glmdir and rh.my_gender_age.glmdir. If you do: {{{ ls lh.my_gender_age.glmdir }}} you should see this: {{{ age/ beta.mgh fsgd.X.mat pca-eres/ rvar.mgh y.fsgd ar1.mgh eres.mgh mri_glmfit.log rstd.mgh Xg.dat }}} and if you do: {{{ ls lh.my_gender_age.glmdir/age }}} you should see this: {{{ C.dat F.mgh gamma.mgh maxvox.dat sig.mgh }}} You can view your results by opening the average subject in tksurfer: {{{ tksurfer average lh inflated }}} and loading in your overlay, '''File -> Load Overlay''' and browse to stats/lh.my_gender_age.glmdir/age/sig.mgh. It should look like this: {{attachment:sig.mgh.jpg}} If you would like to also view the scatter plots associated with this you can do so by '''File -> Load Group Descriptor File''' and browse to stats/lh.my_gender_age.glmdir/y.fsgd. Once that file is loaded, a window titled 'Thickness' will appear. Once that appears, then click on any point on the surface, and the thickness data for that surface vertex will appear in the Thickness plot window. The surface areas in blue indicating cortical thinning with age, as shown in the plot. |
Group Analysis command lines
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
In order to compute the contrasts you will first need to change to the tutorial data directory and setup SUBJECTS_DIR:
cd $TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial
setenv SUBJECTS_DIR ${PWD}Change into the directory you ran the estimation step in, most likely called 'stats':
cd stats
Upon successful completion of the make_average_subject command you should have an average subject in your SUBJECTS_DIR. After successfully completing Exercise A. and Exercise B. you should have an FSGD file called my_gender_age_fsgd.txt and a contrast file called age.mat, both in your SUBJECTS_DIR/stats directory. Confirm that those files exist:
ls my_gender_age_fsgd.txt age.mat
If my_gender_age_fsgd.txt does not exist, complete Exercise A. If age.mat does not exist, complete Exercise B., or just create a file called age.mat containing this line:
0 0 1
The following are sample commands, that can be used with the data, to complete a group analysis:
# For the left hemisphere mris_preproc --fsgd my_gender_age_fsgd.txt \ --target average \ --hemi lh \ --meas thickness \ --out lh.my_gender_age.thickness.mgh
Once this completes you should see the file lh.my_gender_age.thickness.mgh. The next step, the smoothing step, will use this as input:
mri_surf2surf --hemi lh \ --s average \ --sval lh.my_gender_age.thickness.mgh \ --fwhm 10 \ --tval lh.my_gender_age.thickness.10.mgh
Once this is complete you should see the file lh.my_gender_age.thickness.10.mgh. The next step will test the hypothesis you've set up using your contrast vector:
mri_glmfit --y lh.my_gender_age.thickness.10.mgh \ --fsgd my_gender_age_fsgd.txt doss \ --glmdir lh.my_gender_age.glmdir \ --pca \ --surf average lh \ --C age.mat
You should have two new directories in $TUTORIAL_DATA/buckner_data/tutorial_subjs/group_analysis_tutorial/stats - lh.my_gender_age.glmdir and rh.my_gender_age.glmdir. If you do:
ls lh.my_gender_age.glmdir
you should see this:
age/ beta.mgh fsgd.X.mat pca-eres/ rvar.mgh y.fsgd ar1.mgh eres.mgh mri_glmfit.log rstd.mgh Xg.dat
and if you do:
ls lh.my_gender_age.glmdir/age
you should see this:
C.dat F.mgh gamma.mgh maxvox.dat sig.mgh
You can view your results by opening the average subject in tksurfer:
tksurfer average lh inflated
and loading in your overlay, File -> Load Overlay and browse to stats/lh.my_gender_age.glmdir/age/sig.mgh.
It should look like this: 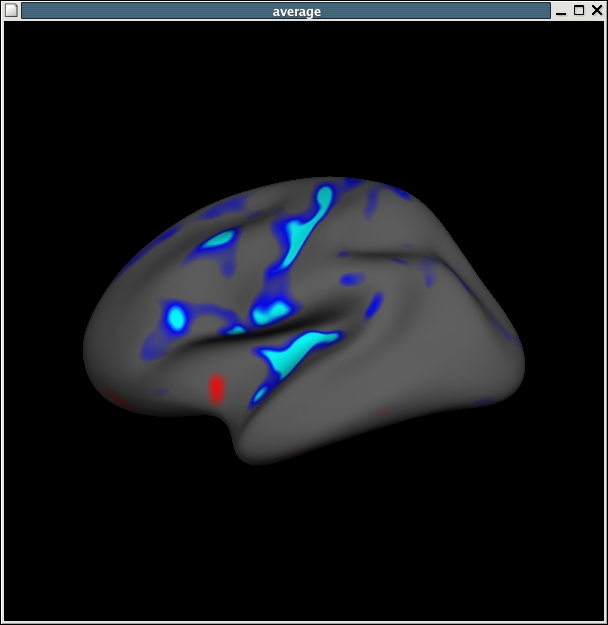
If you would like to also view the scatter plots associated with this you can do so by File -> Load Group Descriptor File and browse to stats/lh.my_gender_age.glmdir/y.fsgd. Once that file is loaded, a window titled 'Thickness' will appear. Once that appears, then click on any point on the surface, and the thickness data for that surface vertex will appear in the Thickness plot window. The surface areas in blue indicating cortical thinning with age, as shown in the plot.
The commands are the same for the right hemisphere, replacing every lh with an rh.
