|
Size: 10938
Comment:
|
Size: 12251
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| [[FsTutorial|Back to Top Tutorial Page]] <<BR>> [[FsTutorial/MultiModal_freeview|Back to Multimodal Top]]<<BR>> The purpose of this tutorial is to get you acquainted with the concepts need to perform fMRI integration in FreeSurfer by interacting with the data from several subjects in a group analysis. You will not learn how to perform fMRI analysis here; that knowledge is already assumed. This tutorial also does not assume any particular directory structure (as would happen in FS-FAST). This tutorial makes use of data from the Functional Biomedical Informatics Research Network (fBIRN, [[http://www.nbirn.net|www.nbirn.net]]).<<BR>> Other multimodal tutorials: [[FsTutorial/MultiModalRegistration_freeview|A. Multimodal Registration]], [[FsTutorial/MultiModalFmriIndividual_freeview|B. Individual fMRI Integration]], [[FsTutorial/MultiModalDtiIndividual_freeview|D. Individual DTI Integration]] <<BR>> <<TableOfContents>> | ## page was renamed from FsTutorial/MultiModalFmriGroup_freeview [[Tutorials|Back to list of all tutorials]]<<BR>>[[FsTutorial|Back to course page]]<<BR>> [[FsTutorial/MultiModal_freeview|Back to Multimodal Tutorial List]] ##Other Multimodal Tutorials: [[FsTutorial/MultiModalDtiIndividual_freeview|B. Individual DTI Integration]], [[FsTutorial/MultiModalFmriIndividual_freeview|C. Individual fMRI Integration]], [[FsTutorial/MultiModalFmriGroup_freeview|D. Surface-based Group fMRI Analysis]] ---- Other Multimodal Tutorials: [[FsTutorial/MultiModalRegistration_freeview|Multimodal Registration]], [[FsTutorial/MultiModalFmriIndividual_freeview|Individual fMRI Integration]], [[FsTutorial/Diffusion_DtiIntegration|Diffusion and DTI Integration]] ---- The purpose of this tutorial is to get you acquainted with the concepts needed to perform fMRI integration in !FreeSurfer by interacting with the data from several subjects in a group analysis. You will not learn how to perform fMRI analysis here; that knowledge is already assumed. This tutorial also does not assume any particular directory structure (as would happen in FS-FAST). This tutorial makes use of data from the Functional Biomedical Informatics Research Network (fBIRN, [[http://www.nbirn.net|www.nbirn.net]]).<<BR>><<BR>> <<TableOfContents(1)>> |
| Line 6: | Line 14: |
| == Preparations == | = Preparations = |
| Line 11: | Line 19: |
| setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs | export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs |
| Line 14: | Line 22: |
| ''To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.'' These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into this directory. You can now skip ahead to the tutorial (below the gray line). | ''To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.'' These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into the directory within the SUBJECTS_DIR that you will use for this part of the tutorial. You can now skip ahead to the tutorial (below the gray line). |
| Line 20: | Line 30: |
| tcsh source your_freesurfer_dir/SetUpFreeSurfer.csh setenv SUBJECTS_DIR $TUTORIAL_DATA/buckner_data/tutorial_subjs |
<source_freesurfer> export TUTORIAL_DATA=<path_to_your_tutorial_data> export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs |
| Line 25: | Line 35: |
| Notice the command to open tcsh. If you are already running the tcsh command shell, then the 'tcsh' command is not necessary. If you are not using the tutorial data you should set your {{{SUBJECTS_DIR}}} to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located. | If you are not using the tutorial data you should set your {{{SUBJECTS_DIR}}} to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located. |
| Line 28: | Line 38: |
| There are five subjects from the fBIRN Phase I acquisition. They are fbirn-10?, where "?" is 1, 3, 4, 5, 6 (note that #2 is missing). Each has a FreeSurfer reconstruction by the name fbirn-anat-10?.v4. The [[FsTutorial/MultiModalRegistration|individual registration]] [[FsTutorial/MultiModalFmriIndividual|ROI Analysis]] have already been performed on all subjects. The data are the results from a sensorimotor paradigm (flashing checkerboard, audible tone, and finger tapping). The raw fMRI data were motion corrected but not smoothed. Each subject has four volumes: | There are five subjects from the fBIRN Phase I acquisition. They are fbirn-10?, where "?" is 1, 3, 4, 5, 6 (note that #2 is missing). Each has a !FreeSurfer reconstruction by the name fbirn-anat-10?.v4. The [[FsTutorial/MultiModalRegistration_freeview|individual registration]] [[FsTutorial/MultiModalFmriIndividual_freeview|ROI Analysis]] have already been performed on all subjects. The data are the results from a sensorimotor paradigm (flashing checkerboard, audible tone, and finger tapping). The raw fMRI data were motion corrected but not smoothed. Each subject has four volumes: ||template.nii ||motion correction template || ||ces.nii ||contrast effect size || ||cesvar.nii ||variance of contrast effect size || ||sig.nii ||signed significance of contrast (-log10(p)) || The contrast is the contrast between the ON and the OFF of the stimulus paradigm (ie, a comparison against baseline). The sig.nii volume has signed -log10(p) values. So, if the p-value = .01, -log10(p) = 2. If the contrast was positive, then the value would be +2, if negative (ie, ON<OFF), then the value would be -2. = Check Registration = First (and always), check the registration (see the [[FsTutorial/MultiModalRegistration_freeview|Registration Tutorial]] for more information). This registration should already be good, so there is no need to make any modifications. In a real analysis, you should check the registrations for all subjects, but that is not necessary here. '''Option A''': Using !FreeView |
| Line 31: | Line 55: |
| template.nii - motion correction template ces.nii - contrast effect size cesvar.nii - variance of contrast effect size sig.nii - signed significance of contrast (-log10(p)) |
freeview -v $SUBJECTS_DIR/fbirn-anat-101.v4/mri/orig.mgz \ fbirn-101/template.nii:reg=fbirn-101/bb.register.lta -f \ $SUBJECTS_DIR/fbirn-anat-101.v4/surf/lh.white:edgecolor=green \ $SUBJECTS_DIR/fbirn-anat-101.v4/surf/rh.white:edgecolor=green \ -viewport coronal |
| Line 36: | Line 61: |
| The contrast is the contrast between the ON and the OFF (ie, a comparison against baseline). The sig.nii volume has signed -log10(p) values. So, if the p-value = .01, -log10(p) = 2. If the contrast was positive, then the value would be +2, if negative (ie, ON<OFF), then the value would be -2. | Use Alt+v to toggle between the template and the orig, using the surfaces as a guide to make sure they are aligned. |
| Line 38: | Line 63: |
| = Check Registration = First (and always), check the registration (see the [[FsTutorial/MulitModalRegistration|Registration Tutorial]] for more information). |
'''Option B''': Using tkregister2 |
| Line 41: | Line 65: |
| ---- | |
| Line 44: | Line 67: |
| --reg fbirn-101/bb.register.dat --surf | --reg fbirn-101/bb.register.lta --surf |
| Line 46: | Line 69: |
| ---- This registration should already be good, so there is no need to make any modifications. In a real analysis, you should check the registrations for all subjects, but that is not necessary here. = Surface-based Group fMRI Analysis = |
== Surface-based Group fMRI Analysis == |
| Line 57: | Line 77: |
| Only the first step is different than an anatomical (eg, thickness) analysis. Still, you run mri_preproc, but with different arguments: | Only the first step is different than an anatomical (eg, thickness) analysis. Still, you run mris_preproc, but with different arguments: |
| Line 59: | Line 79: |
| ---- | |
| Line 62: | Line 81: |
| --iv fbirn-101/ces.nii fbirn-101/bb.register.dat \ --iv fbirn-103/ces.nii fbirn-103/bb.register.dat \ --iv fbirn-104/ces.nii fbirn-104/bb.register.dat \ --iv fbirn-105/ces.nii fbirn-105/bb.register.dat \ --iv fbirn-106/ces.nii fbirn-106/bb.register.dat \ |
--iv fbirn-101/ces.nii fbirn-101/bb.register.lta \ --iv fbirn-103/ces.nii fbirn-103/bb.register.lta \ --iv fbirn-104/ces.nii fbirn-104/bb.register.lta \ --iv fbirn-105/ces.nii fbirn-105/bb.register.lta \ --iv fbirn-106/ces.nii fbirn-106/bb.register.lta \ |
| Line 70: | Line 89: |
| ---- | |
| Line 73: | Line 91: |
| 1. The target surface is the left hemisphere of fsaverage 1. Each input volume and corresponding registration is listed separately |
1. The target surface is the left hemisphere of fsaverage. 1. Each input volume and corresponding registration is listed separately. |
| Line 77: | Line 95: |
| The output is lh.ces.mgh. It has the same size as any other surface overlay for fsaverage eg, lh.thickness. To see it's dimensions, run: | The output is lh.ces.mgh. It has the same size as any other surface overlay for fsaverage eg, lh.thickness. To see its dimensions, run: |
| Line 79: | Line 97: |
| ---- | |
| Line 83: | Line 100: |
| ---- | |
| Line 87: | Line 103: |
| ---- | |
| Line 92: | Line 107: |
| ---- | |
| Line 95: | Line 109: |
| 1. Full-Width/Half-Max (fwhm) is 5 mm 1. The smoothing is only perform in regions of legitimate cortex (--cortex, which uses the lh.cortex.label). |
1. Full-Width/Half-Max (fwhm) is 5 mm. 1. The smoothing is only performed in regions of legitimate cortex (--cortex, which uses the lh.cortex.label). |
| Line 101: | Line 115: |
| ---- | |
| Line 106: | Line 119: |
| ---- ---- |
|
| Line 109: | Line 120: |
| freeview -f $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:overlay=lh.ces.sm05.osgm/osgm/sig.mgh:overlay_threshold=2,5 | freeview -f \ $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:annot_outline=1:overlay=lh.ces.sm05.osgm/osgm/sig.mgh:overlay_threshold=2,5 \ -viewport 3d |
| Line 111: | Line 124: |
| ---- Once the aparc is loaded, click the '''Show outline only''' checkbox to see the significance map. {{attachment:glmfit_fv.jpg||height="370",width="518"}} {{attachment:glmfitmed_fv.jpg||height="370",width="553"}} |
{{attachment:glmfit_fv.jpg||height="370",width="518"}} {{attachment:glmfitmed_fv.jpg||height="369",width="553"}} |
| Line 119: | Line 129: |
| The purpose of this tutorial is to combine individual stats files into one summary table file that can be loaded into a spreadsheet program such as excel or oocalc. The individual ROI analyses were performed as described in the [[FsTutorial/MulitModalFmriIndividual|Individual fMRI Integration Tutorial]]. Namely, stats files were created by mri_segstats using no functional contraint, an unsigned (abs) constraint, and a positive (pos) constraint to create stats files in each directory. Furthermore, only 4 segmentations were summarized: | The purpose of this tutorial is to combine individual stats files into one summary table file. The table files can be opened with any text editor, or also loaded into a spreadsheet program such as MS Excel or !OpenOffice Calc if you have one installed. The individual ROI analyses were performed as described in the [[FsTutorial/MultiModalFmriIndividual_freeview|Individual fMRI Integration Tutorial]]. Namely, stats files were created by mri_segstats using no functional contraint, an unsigned (abs) constraint, and a positive (pos) constraint to create stats files in each directory. Furthermore, only 4 segmentations were summarized: ||1021 ||ctx-lh-pericalcarine || ||1022 ||ctx-lh-postcentral || ||1030 ||ctx-lh-superiortemporal || ||17 ||Left-Hippocampus || |
| Line 121: | Line 135: |
| 1. 1021=ctx-lh-pericalcarine 1. 1022=ctx-lh-postcentral 1. 1030=ctx-lh-superiortemporal 1. 17=Left-Hippocampus |
|
| Line 127: | Line 139: |
| ---- | |
| Line 138: | Line 149: |
| ---- Click [[FsTutorial/MultiModalFmriIndividual/BbStats#head-ddb7dd6be9b64b6a490cc20e619cb715e59f4721|here]] to see the output (ces.pos-masked.vol.stats). Notes: |
Click [[FsTutorial/MultiModalFmriIndividual/BbStats#ces.pos-masked.vol.stats|here]] to see the output (ces.pos-masked.vol.stats). Notes: |
| Line 141: | Line 151: |
| 1. The values are the volume in mm3 of the ON>OFF voxels whose pvalues < .01. 1. The value first column is just the row number (starting from 0). Normally, the subject name would be here, but the subject name is not input to the command above. |
1. The values are the volume in mm3 of the ON>OFF voxels whose p-values < .01. 1. The value in the first column is just the row number (starting from 0). Normally, the subject name would be here, but the subject name was not specified as input in the above command. |
| Line 145: | Line 155: |
| ---- | |
| Line 156: | Line 165: |
| ---- Click [[FsTutorial/MultiModalFmriIndividual/BbStats#head-ddb7dd6be9b64b6a490cc20e619cb715e59f4721|here]] to see the output (ces.abs-masked.mean.stats). Notes: |
Click [[FsTutorial/MultiModalFmriIndividual/BbStats#ces.abs-masked.mean.stats|here]] to see the output (ces.abs-masked.mean.stats). Notes: |
| Line 159: | Line 167: |
| 1. The values are the mean HRF values (raw MR units) of the ON>OFF or ON<OFF voxels whose pvalues < .01. | 1. The values are the mean HRF values (raw MR units) of the ON>OFF or ON<OFF voxels whose p-values < .01. |
| Line 163: | Line 171: |
| == Measure Mean HRF Contrast of Postiive Activation in Each ROI == ---- |
== Measure Mean HRF Contrast of Positive Activation in Each ROI == |
| Line 175: | Line 182: |
| ---- Click [[FsTutorial/MultiModalFmriIndividual/BbStats#head-ddb7dd6be9b64b6a490cc20e619cb715e59f4721|here]] to see the output (ces.pos-masked.mean.stats). Notes: |
Click [[FsTutorial/MultiModalFmriIndividual/BbStats#ces.pos-masked.mean.stats|here]] to see the output (ces.pos-masked.mean.stats). Notes: |
| Line 178: | Line 184: |
| 1. The values are the mean HRF values (raw MR units) of the ON>OFF voxels whose pvalues < .01. | 1. The values are the mean HRF values (raw MR units) of the ON>OFF voxels whose p-values < .01. |
| Line 182: | Line 188: |
| Other multimodal tutorials: [[FsTutorial/MultiModalRegistration_freeview|A. Multimodal Registration]], [[FsTutorial/MultiModalFmriIndividual_freeview|B. Individual fMRI Integration]], [[FsTutorial/MultiModalDtiIndividual_freeview|D. Individual DTI Integration]] <<BR>> | ## Other multimodal tutorials: [[FsTutorial/MultiModalRegistration_freeview|A. Multimodal Registration]], [[FsTutorial/MultiModalDtiIndividual_freeview|B. Individual DTI Integration]], [[FsTutorial/MultiModalFmriIndividual_freeview|C. Individual fMRI Integration]] ---- Other Multimodal Tutorials: [[FsTutorial/MultiModalRegistration_freeview|Multimodal Registration]], [[FsTutorial/MultiModalFmriIndividual_freeview|Individual fMRI Integration]], [[FsTutorial/Diffusion_DtiIntegration|Diffusion and DTI Integration]]<<BR>> ---- [[https://surfer.nmr.mgh.harvard.edu/fswiki/Tutorials|Back to list of all tutorials]]<<BR>>[[https://surfer.nmr.mgh.harvard.edu/fswiki/FsTutorial|Back to course page]]<<BR>> [[FsTutorial/MultiModal_freeview|Back to Multimodal Tutorial List]] |
Back to list of all tutorials
Back to course page
Back to Multimodal Tutorial List
Other Multimodal Tutorials: Multimodal Registration, Individual fMRI Integration, Diffusion and DTI Integration
The purpose of this tutorial is to get you acquainted with the concepts needed to perform fMRI integration in FreeSurfer by interacting with the data from several subjects in a group analysis. You will not learn how to perform fMRI analysis here; that knowledge is already assumed. This tutorial also does not assume any particular directory structure (as would happen in FS-FAST). This tutorial makes use of data from the Functional Biomedical Informatics Research Network (fBIRN, www.nbirn.net).
fMRI Basics
In fMRI, stimuli are presented to a subject, which creates a BOLD hemodynamic response function (HRF) in certain areas of the brain. The analysis is performed by first performing motion correction, then correlating each voxel's time course with the stimulus schedule convolved with an assumed HRF shape. The result is an estimate of the HRF amplitude for each condition at each voxel, contrasts of the HRF amplitudes of various conditions, the variance of this contrast, and some measure of the significance (eg, p, t, F, or z) map. All these maps are aligned with the motion correction template, which should be used as the registration template.
Preparations
If You're at an Organized Course
If you are taking one of the formally organized courses, everything has been set up for you on the provided laptop. The only thing you will need to do is run the following commands in every new terminal window (aka shell) you open throughout this tutorial. Copy and paste the commands below to get started:
export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR/multimodal/fmri
To copy: Highlight the command in the box above, right click and select copy (or use keyboard shortcut Ctrl+c), then use the middle button of your mouse to click inside the terminal window (this will paste the command). Press enter to run the command.
These two commands set the SUBJECTS_DIR variable to the directory where the data is stored and then navigates into the directory within the SUBJECTS_DIR that you will use for this part of the tutorial. You can now skip ahead to the tutorial (below the gray line).
If You're not at an Organized Course
If you are NOT taking one of the formally organized courses, then to follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names. These are the commands that you need to run before getting started:
<source_freesurfer> export TUTORIAL_DATA=<path_to_your_tutorial_data> export SUBJECTS_DIR=$TUTORIAL_DATA/buckner_data/tutorial_subjs cd $SUBJECTS_DIR/multimodal/fmri
If you are not using the tutorial data you should set your SUBJECTS_DIR to the directory in which the recon(s) of the subject(s) you will use for this tutorial are located.
There are five subjects from the fBIRN Phase I acquisition. They are fbirn-10?, where "?" is 1, 3, 4, 5, 6 (note that #2 is missing). Each has a FreeSurfer reconstruction by the name fbirn-anat-10?.v4. The individual registration ROI Analysis have already been performed on all subjects. The data are the results from a sensorimotor paradigm (flashing checkerboard, audible tone, and finger tapping). The raw fMRI data were motion corrected but not smoothed. Each subject has four volumes:
template.nii |
motion correction template |
ces.nii |
contrast effect size |
cesvar.nii |
variance of contrast effect size |
sig.nii |
signed significance of contrast (-log10(p)) |
The contrast is the contrast between the ON and the OFF of the stimulus paradigm (ie, a comparison against baseline). The sig.nii volume has signed -log10(p) values. So, if the p-value = .01, -log10(p) = 2. If the contrast was positive, then the value would be +2, if negative (ie, ON<OFF), then the value would be -2.
Check Registration
First (and always), check the registration (see the Registration Tutorial for more information). This registration should already be good, so there is no need to make any modifications. In a real analysis, you should check the registrations for all subjects, but that is not necessary here.
Option A: Using FreeView
freeview -v $SUBJECTS_DIR/fbirn-anat-101.v4/mri/orig.mgz \ fbirn-101/template.nii:reg=fbirn-101/bb.register.lta -f \ $SUBJECTS_DIR/fbirn-anat-101.v4/surf/lh.white:edgecolor=green \ $SUBJECTS_DIR/fbirn-anat-101.v4/surf/rh.white:edgecolor=green \ -viewport coronal
Use Alt+v to toggle between the template and the orig, using the surfaces as a guide to make sure they are aligned.
Option B: Using tkregister2
tkregister2 --mov fbirn-101/template.nii \ --reg fbirn-101/bb.register.lta --surf
Surface-based Group fMRI Analysis
The group analysis is done in three stages:
- Assemble data
- Spatially smooth data on surface (if desired)
- GLM Analysis
Assemble Data
Only the first step is different than an anatomical (eg, thickness) analysis. Still, you run mris_preproc, but with different arguments:
mris_preproc --target fsaverage --hemi lh \ --iv fbirn-101/ces.nii fbirn-101/bb.register.lta \ --iv fbirn-103/ces.nii fbirn-103/bb.register.lta \ --iv fbirn-104/ces.nii fbirn-104/bb.register.lta \ --iv fbirn-105/ces.nii fbirn-105/bb.register.lta \ --iv fbirn-106/ces.nii fbirn-106/bb.register.lta \ --projfrac 0.5 \ --out lh.ces.mgh
Notes:
- The target surface is the left hemisphere of fsaverage.
- Each input volume and corresponding registration is listed separately.
- The fMRI data are sampled half-way between the white and pial surfaces ("--projfrac 0.5").
The output is lh.ces.mgh. It has the same size as any other surface overlay for fsaverage eg, lh.thickness. To see its dimensions, run:
mri_info lh.ces.mgh
You will see "dimensions: 163842 x 1 x 1 x 5", indicating that there are 163842 "columns" (ie, vertices), 1 "row", and 1 "slice". There are 5 frames, one for each subject.
Surface Smoothing
mri_surf2surf --hemi lh --s fsaverage --fwhm 5 --cortex\ --sval lh.ces.mgh --tval lh.ces.sm05.mgh
Notes:
- Full-Width/Half-Max (fwhm) is 5 mm.
- The smoothing is only performed in regions of legitimate cortex (--cortex, which uses the lh.cortex.label).
GLM Fit
This performs a one-sample, group mean (OSGM) just looking for places where the CES is different than 0.
mri_glmfit --y lh.ces.sm05.mgh --surf fsaverage lh \ --osgm --glmdir lh.ces.sm05.osgm --cortex
freeview -f \ $SUBJECTS_DIR/fsaverage/surf/lh.inflated:annot=aparc.annot:annot_outline=1:overlay=lh.ces.sm05.osgm/osgm/sig.mgh:overlay_threshold=2,5 \ -viewport 3d
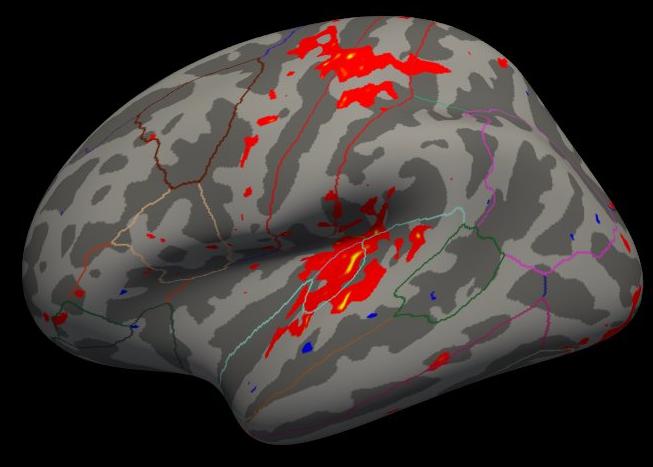
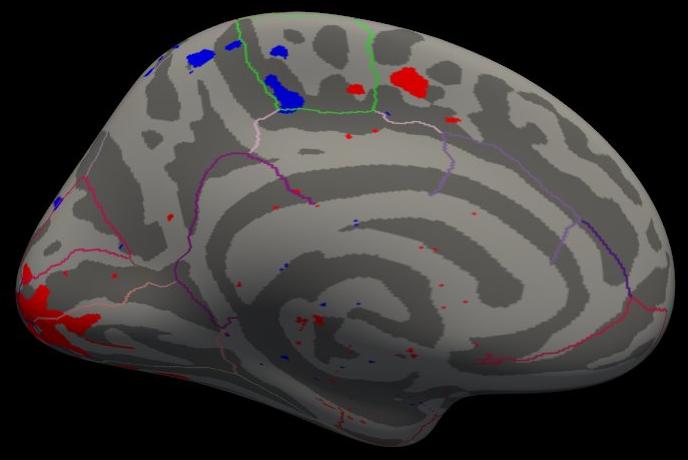
Note that the activation is light because there are only five subjects, but the activation that is there is in the right place.
Group ROI fMRI Analysis
The purpose of this tutorial is to combine individual stats files into one summary table file. The table files can be opened with any text editor, or also loaded into a spreadsheet program such as MS Excel or OpenOffice Calc if you have one installed. The individual ROI analyses were performed as described in the Individual fMRI Integration Tutorial. Namely, stats files were created by mri_segstats using no functional contraint, an unsigned (abs) constraint, and a positive (pos) constraint to create stats files in each directory. Furthermore, only 4 segmentations were summarized:
1021 |
ctx-lh-pericalcarine |
1022 |
ctx-lh-postcentral |
1030 |
ctx-lh-superiortemporal |
17 |
Left-Hippocampus |
Measure Volume of Positive Activation in Each ROI
asegstats2table \ --meas volume \ --tablefile ces.pos-masked.vol.stats \ --i fbirn-101/ces.pos-masked.bb.stats \ fbirn-103/ces.pos-masked.bb.stats \ fbirn-104/ces.pos-masked.bb.stats \ fbirn-105/ces.pos-masked.bb.stats \ fbirn-106/ces.pos-masked.bb.stats
Click here to see the output (ces.pos-masked.vol.stats). Notes:
The values are the volume in mm3 of the ON>OFF voxels whose p-values < .01.
- The value in the first column is just the row number (starting from 0). Normally, the subject name would be here, but the subject name was not specified as input in the above command.
Measure Mean HRF Contrast of Unsigned Activation in Each ROI
asegstats2table \ --meas mean \ --tablefile ces.abs-masked.mean.stats \ --i fbirn-101/ces.abs-masked.bb.stats \ fbirn-103/ces.abs-masked.bb.stats \ fbirn-104/ces.abs-masked.bb.stats \ fbirn-105/ces.abs-masked.bb.stats \ fbirn-106/ces.abs-masked.bb.stats
Click here to see the output (ces.abs-masked.mean.stats). Notes:
The values are the mean HRF values (raw MR units) of the ON>OFF or ON<OFF voxels whose p-values < .01.
- Some of the values are negative. This is possible because the mask is unsigned (abs).
- In particular, the cross-subject average of Left-Hippocampus is near 0. This is expected because the sensorimotor task should not activate hippocampus.
Measure Mean HRF Contrast of Positive Activation in Each ROI
asegstats2table \ --meas mean \ --tablefile ces.pos-masked.mean.stats \ --i fbirn-101/ces.pos-masked.bb.stats \ fbirn-103/ces.pos-masked.bb.stats \ fbirn-104/ces.pos-masked.bb.stats \ fbirn-105/ces.pos-masked.bb.stats \ fbirn-106/ces.pos-masked.bb.stats
Click here to see the output (ces.pos-masked.mean.stats). Notes:
The values are the mean HRF values (raw MR units) of the ON>OFF voxels whose p-values < .01.
- NONE of the values are negative. This is because the mask is positive (pos).
In particular, the cross-subject average of Left-Hippocampus is NOT close to 0 because only positive values were chosen. So, one must be careful how to interpret and draw conclusions from these numbers.
Other Multimodal Tutorials: Multimodal Registration, Individual fMRI Integration, Diffusion and DTI Integration
Back to list of all tutorials
Back to course page
Back to Multimodal Tutorial List
