Making Edits to the White Matter
To follow this exercise exactly be sure you've downloaded the tutorial data set before you begin. If you choose not to download the data set you can follow these instructions on your own data, but you will have to substitute your own specific paths and subject names.
Sometimes the white matter is not segmented correctly: sometimes voxels that should be white matter are excluded, and other times voxels that should not be white matter are included in error. Either of these occurrences can be fixed with simple manual edits. Below you will find examples of a few of the common problems and how to fix them. For some people it is easiest to see these problems when looking at the inflated surface in freeview. Others can pick them out while viewing the volumes in freeview. Either method is fine for identifying these problems. However they can only be fixed using freeview with the wm.mgz volume.
Geometric inaccuracy due to brain lesion
wm1_edits_before is an example of white matter being excluded from the wm.mgz volume due to the presence of a brain lesion. This page will walk you through fixing this specific example as well as other common problems with the wm.mgz volume. First, make sure you have wm1_edits_before loaded in freeview (you may have already run this command):
freeview -v wm1_edits_before/mri/brainmask.mgz \ wm1_edits_before/mri/wm.mgz:colormap=heat:opacity=0.4 \ -f wm1_edits_before/surf/lh.white:edgecolor=blue \ wm1_edits_before/surf/lh.pial:edgecolor=red \ wm1_edits_before/surf/rh.white:edgecolor=blue \ wm1_edits_before/surf/rh.pial:edgecolor=red \ wm1_edits_before/surf/rh.inflated:visible=0 \ wm1_edits_before/surf/lh.inflated:visible=0
Again, this will bring up the brainmask.mgz volume, the wm.mgz volume, and the surfaces for both hemispheres. Make sure the wm.mgz is on top of the brainmask.mgz and highlighted and then click in the main viewing area and use Alt-V to toggle the wm.mgz on and off to see the brain underneath. You may also use Alt-C or the arrows  to flip between the wm.mgz, brainmask.mgz, and T1.mgz if you have more than two volumes loaded. With the wm.mgz loaded on top of the brainmask.mgz volume, it should look like this (below). For a reminder on why we use the heat overlay, please refer to the previous exercise
to flip between the wm.mgz, brainmask.mgz, and T1.mgz if you have more than two volumes loaded. With the wm.mgz loaded on top of the brainmask.mgz volume, it should look like this (below). For a reminder on why we use the heat overlay, please refer to the previous exercise
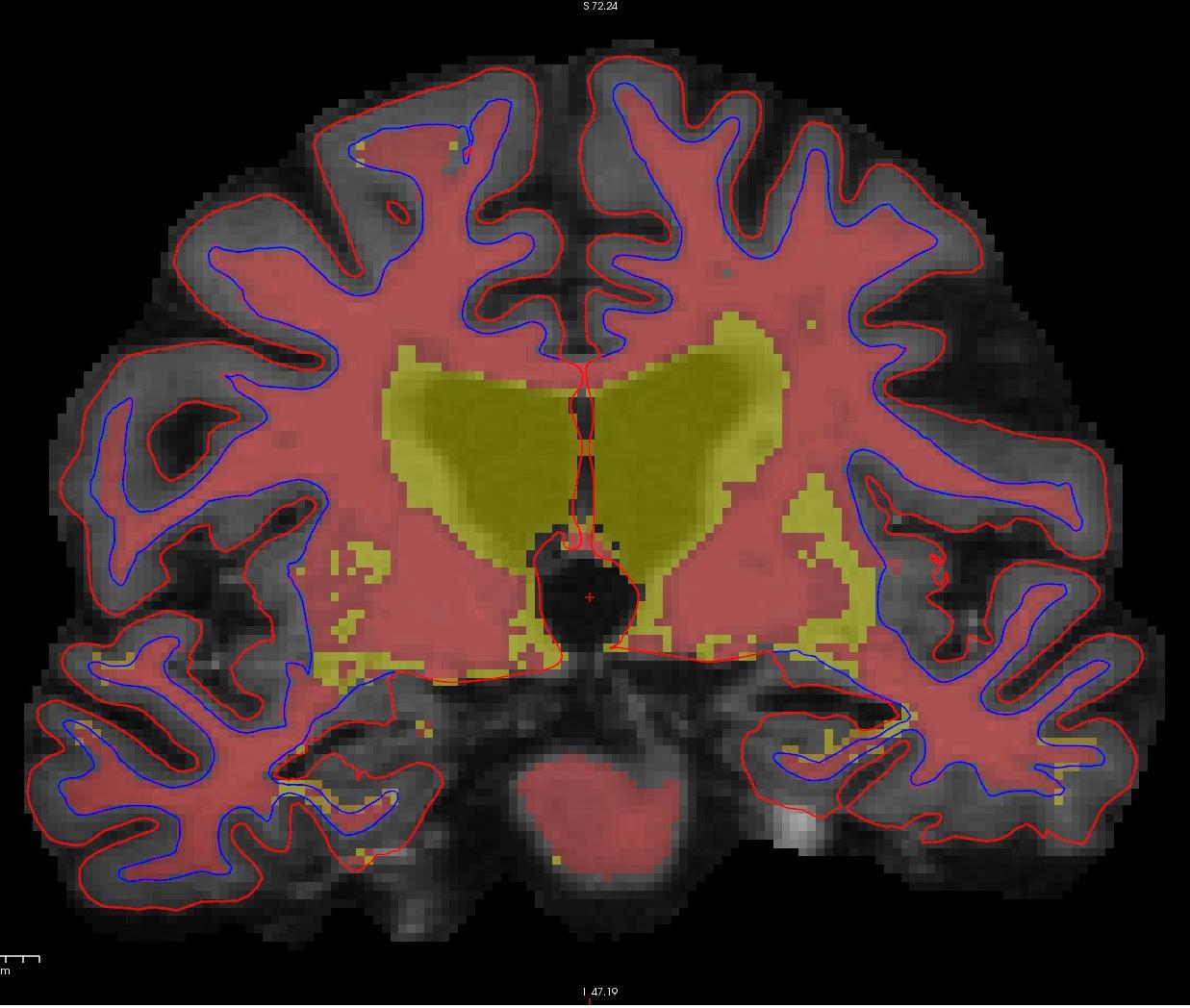
If you look at coronal slice 159 you will notice an area on the right hemisphere where the white surface (blue line) does not follow the surface of the brain, but in fact cuts into it. This geometric inaccuracy is caused by a lesion where white matter has been marked as non-white matter.
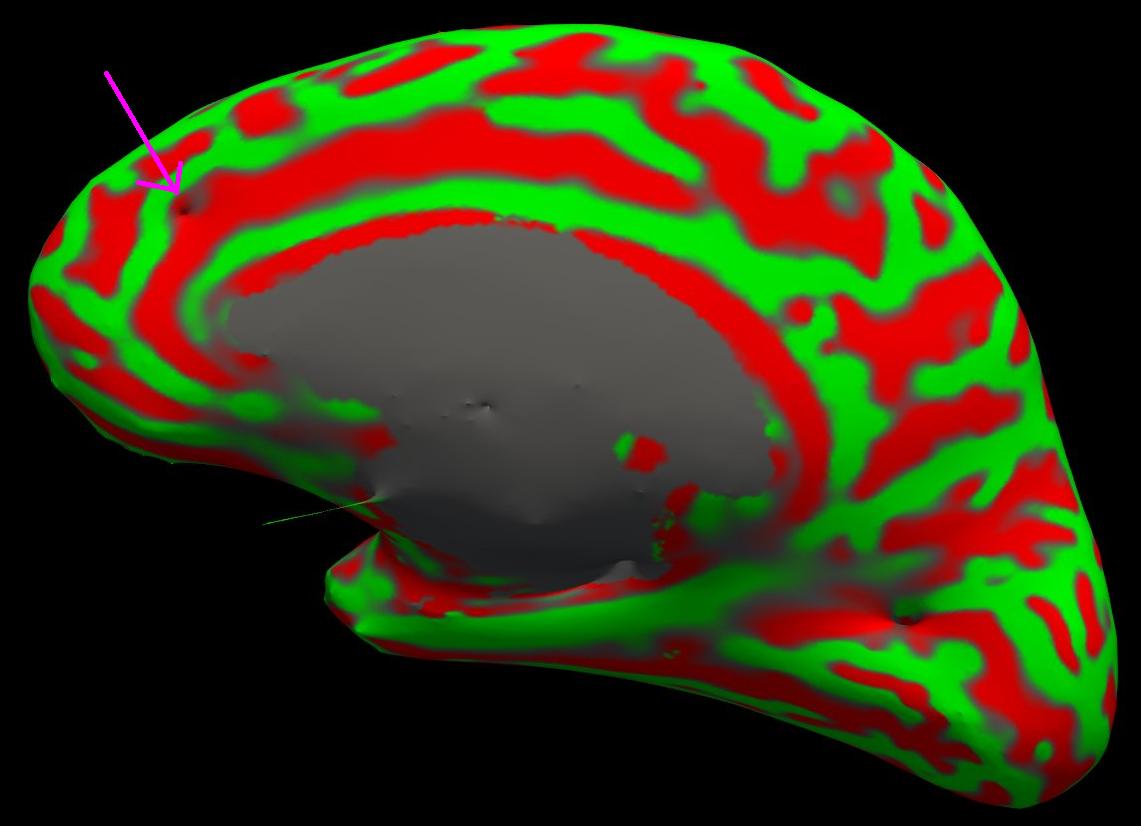
You can see this as a dimple or hole on the inflated surface in the 3D view as well (see next image). Switch to the 3D view and check the box next to the rh.inflated surface. You may want to uncheck the other surfaces. If your view of the surfaces is obstructed by the 2D slices appearing in the 3D view, you can shut them off by pressing Ctrl-Shft-S on the keyboard or by clicking the View option in Freesurfer and then clicking on Show Slices (3D View). You can see the dimple or hole on the medial side.
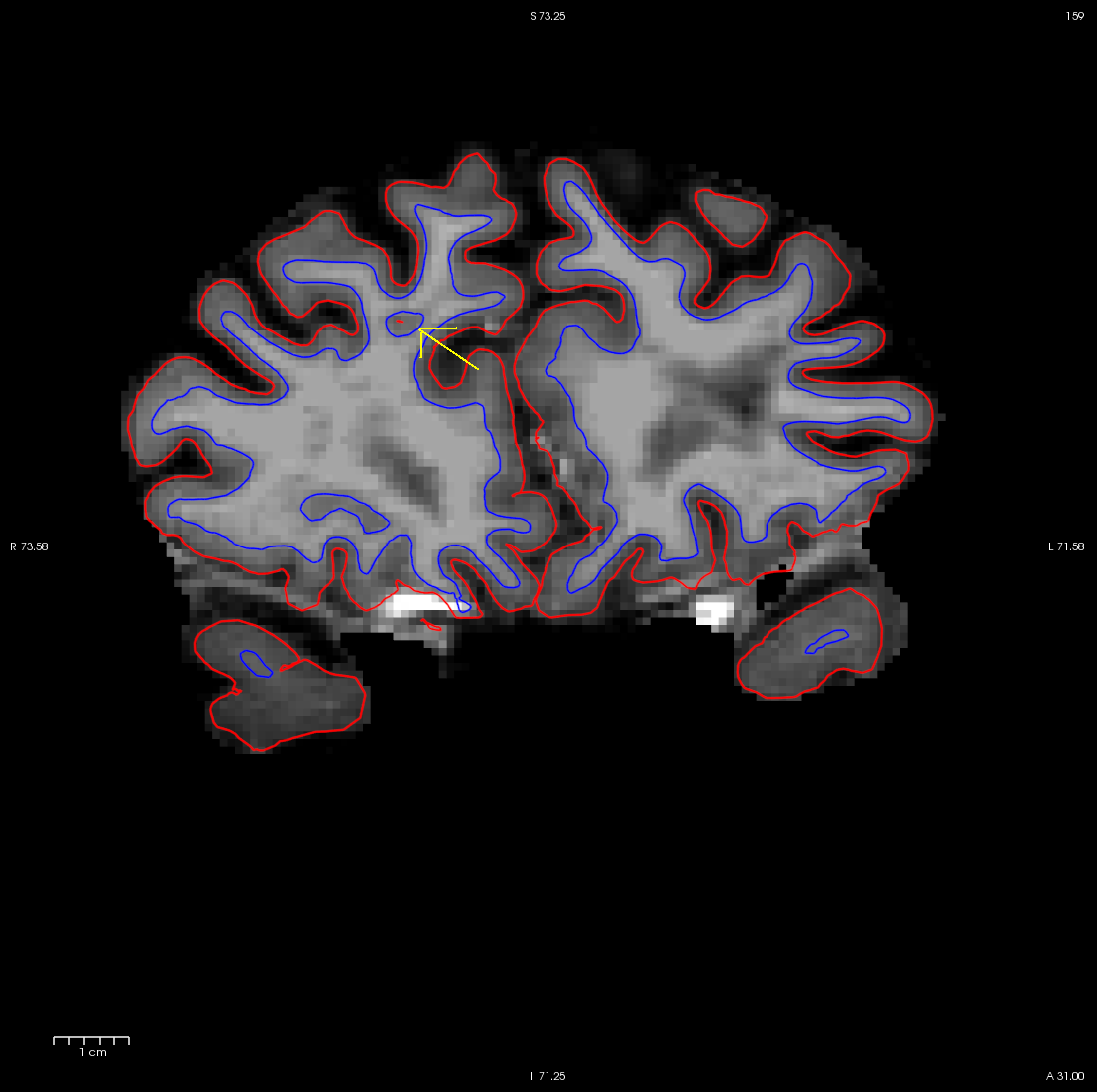
With the wm.mgz volume loaded into freeview you can see that this area has been left out of the wm volume completely (coronal slice 159).
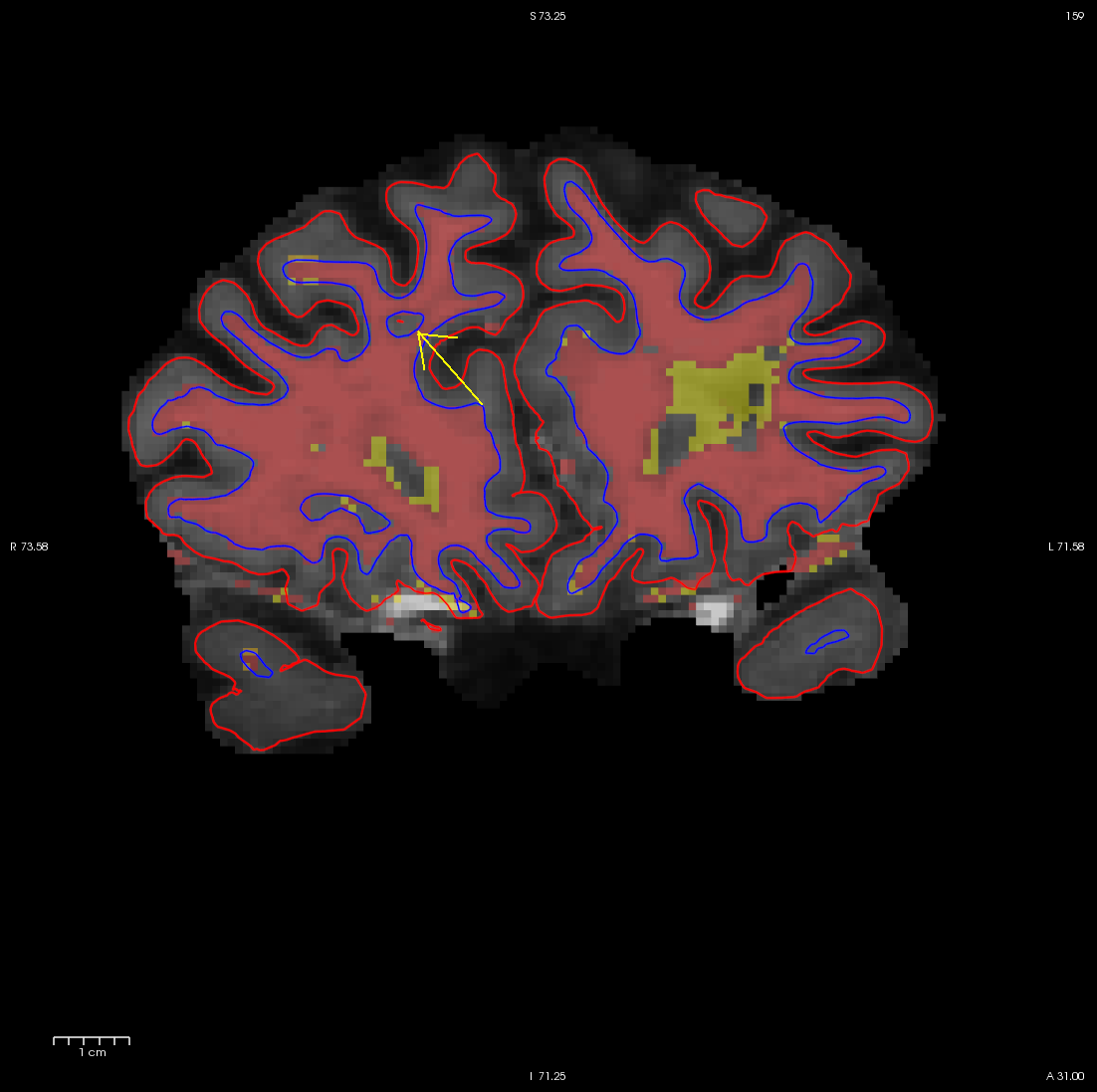
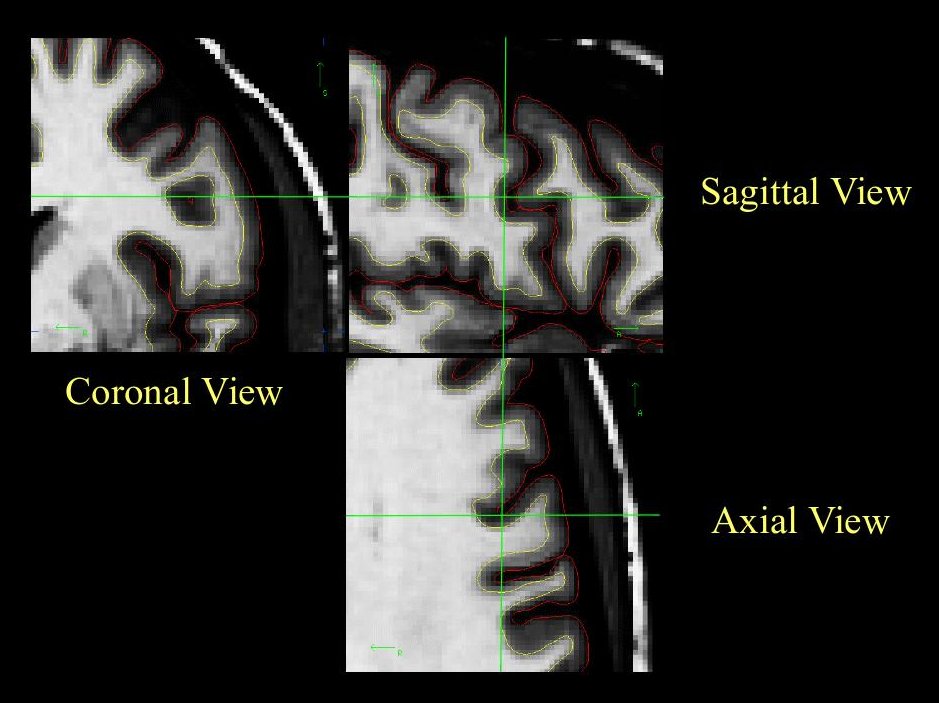
It is always a good idea to look at this area in all three views, to make sure that the hole you see is indeed a segmentation error (it can be very difficult to determine the shape of a 3D object in a 2D slice). In the image below, what appears to be a hole is actually just a sulcus.
To fix this problem you will need to fill in the missing voxels in the wm.mgz volume. First, it's a good idea to use the page up and down keys (or the up and down arrows) to scroll through the individual slices in the volume until you get a good idea of where the problem starts and ends. You will want to start filling in voxels when the inaccuracy appears, and keep filling them in slice by slice until the problem is no longer visible. The coronal view and a brush radius of one or two are good settings for painting in voxels. Zoom in and out either by using the scroll button on your mouse. To begin editing voxels, make sure the wm.mgz is highlighted in the volumes list, and is on top of the brainmask.mgz. Click on the Recon Edit option (  ) and enter a brush size of either 1 or 2. You will see that the recon editing checkbox has set the brush value to 255 and the eraser value to 1.
) and enter a brush size of either 1 or 2. You will see that the recon editing checkbox has set the brush value to 255 and the eraser value to 1.
Remember: the wm.mgz must be on top and the 'Recon editing' box checked (brush value set to 255) in order to edit voxels in the wm.mgz, if it is not visible on top you will be editing the brainmask.mgz. You should only be editing the brainmask for pial edits and not wm edits.
You are now ready to edit. Use your left mouse button fill in the wm hole with voxels. If you fill in too many, you can press Shift while using your left mouse button to erase voxels. You can keep flipping back between the brainmask and the wm volume using your Alt-V or Alt-C combination if you want to check that what you are filling in is indeed white matter. At any time you can use the Crtl-Z combination to undo as many actions as you like.
Video Tutorial:
While filling the hole in slice 160, you will notice that the hole is not completely enclosed. To see where the boundary should be, switch back and forth between the brainmask.mgz and wm.mgz volume with Alt-C. Use the brainmask.mgz volume as a guide, and make the boundary of the voxels that you paint in correspond as closely as possible to the boundaries suggested in the brain volume. Go to the next slice, and fill in this slice in the same way. You will need to continue filling in slices in this fashion until the region is completely filled in. If you would like to compare your edits to ones that have been done for you you can open a second freeview window with the wm1_edits_after wm.mgz volume, and for each slice check your results with the corresponding slice in the wm1_edits_after volume. At any time, you can save the changes you've made by clicking on the wm.mgz (for the sake of the course, do not press save), making sure it is highlighted in the menu to the left side of the viewing area and click the save volume button  or save in the file menu. You can verify your results by viewing the changes made to the wm1_edits_after wm.mgz volume to view this corrected subject (which was corrected by following the detailed instructions provided to you):
or save in the file menu. You can verify your results by viewing the changes made to the wm1_edits_after wm.mgz volume to view this corrected subject (which was corrected by following the detailed instructions provided to you):
freeview -v wm1_edits_after/mri/brainmask.mgz \ wm1_edits_after/mri/wm.mgz:colormap=heat:opacity=0.4 \ -f wm1_edits_after/surf/lh.white:edgecolor=blue \ wm1_edits_after/surf/lh.pial:edgecolor=red \ wm1_edits_after/surf/rh.white:edgecolor=blue \ wm1_edits_after/surf/rh.pial:edgecolor=red \ wm1_edits_after/surf/rh.inflated:visible=0 \ wm1_edits_after/surf/lh.inflated:visible=0
Tips:
- Sometimes regions may appear enclosed in some slices (i.e. appear as holes), open in subsequent slices (i.e. no longer appear as holes), then enclosed again as you scroll forward. The rule of thumb when editing these is to keep filling until you reach the slice where they finally open up and are no longer enclosed.
- When editing geometric inaccuracies in the future, you may find it easier to edit the first slice and last slice first, thereby 'capping' the slices that need filling. Then you just need to fill all the slices between the endcaps.
- Sometimes there is variation between a subject's left and right hemispheres, so that in a particular slice one hemisphere's region will finally 'open up', but the other hemisphere's region is still enclosed. In such cases, continue to fill the enclosed region only, even though the other is open. This will address the topological problem.
- Problems like the one shown here, due to brain lesions, should always be fixed with edits to the wm.mgz volume. Do not use control points to try and automatically adjust the intensity in these areas.
After you have saved all of your edits, you could recreate the final surfaces with the command:
recon-all -autorecon2-wm -autorecon3 -subjid wm1_edits_before
Do not run this command if you are conducting the tutorial! This step will take a long time and there is no need to run it for the tutorial purposes.
